UT Austin
From 2006.igem.org
| Line 26: | Line 26: | ||
We are looking to build circuits which demonstrate the unique regulatory capabilites of the light dependent system developed by the UT/UCSF team last year. One such instantiation is the original [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5407 Edge Detector] (see below) from last year which we are finally finishing. | We are looking to build circuits which demonstrate the unique regulatory capabilites of the light dependent system developed by the UT/UCSF team last year. One such instantiation is the original [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5407 Edge Detector] (see below) from last year which we are finally finishing. | ||
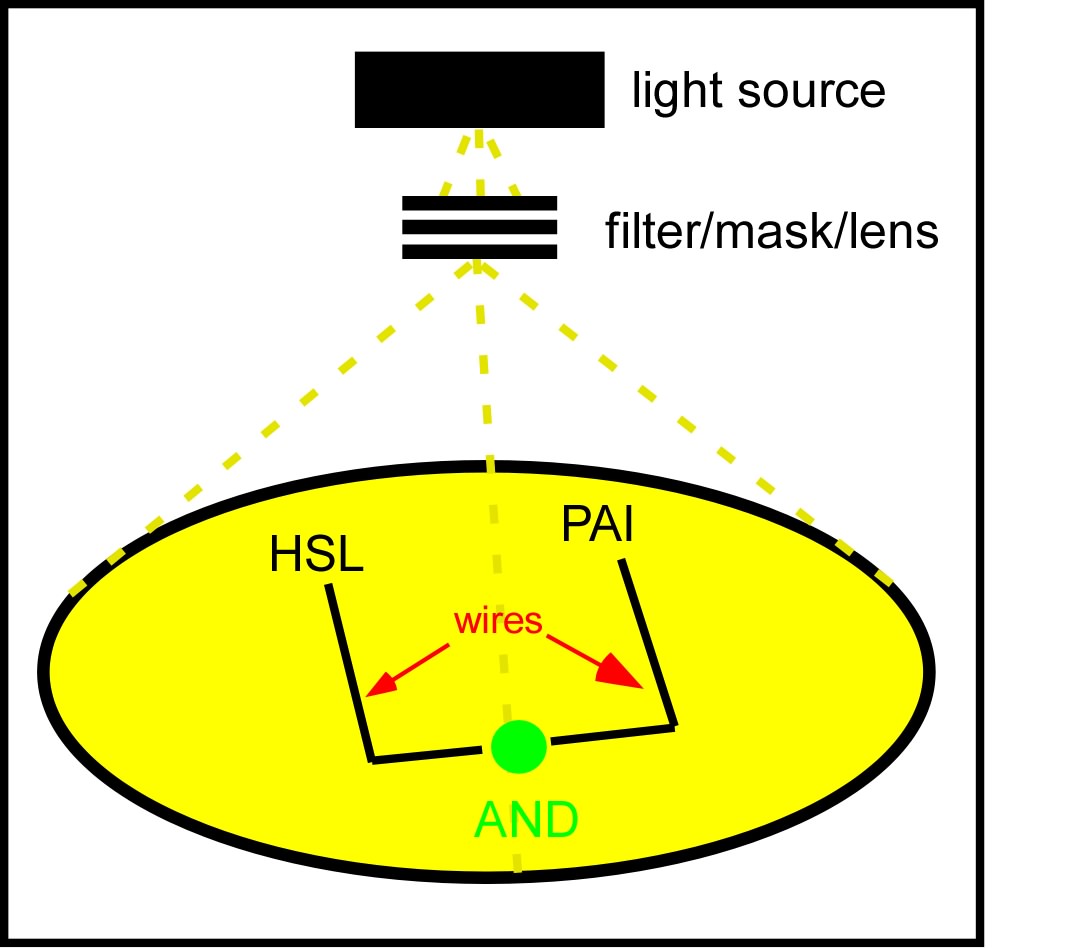
| - | [[Image:light_wires_agar_plate.jpg|thumb|right| | + | [[Image:light_wires_agar_plate.jpg|thumb|right|310px|Light wires. A genetically identical lawn of ''E.coli'' will be transformed into a biochemical circuit board capable of conducting a current of exogenously added quorum sensing compounds (HSL, PAI). At the intersection of two orthogonal biochemical currents, the cells will perform a logical AND operation, signalling the succesful propagation of two compounds.]] |
Revision as of 06:07, 1 November 2005
The UT Austin team, like last year, is extremely informal. We have no classes nor particularly organized meetings. Anyone who wishes to contribute is welcome to do so. Thanks to the natural enthusiasm of our team and occasional whip cracking from Andy we have made some interesting progress.
Last year we planned on building a light activated edge detector. Thanks to the the incredible contribution of an engineered light detector from Anselm Levskaya at [http://www.voigtlab.ucsf.edu/ Voigt Lab UCSF] we were able to take bacterial photographs (publication pending). This was only the first step in our planned goal and in fact we are still working on building the edge detector.
This year we have several project plans which, if they don't self-assemble into green goo and eat the molecular biology building, should be fairly amusing. Being at heart a bunch of hackers, we believe that the greatest contribution to the field will come from actual experiments and thus we are plowing ahead with our experiments while or before modeling our system. Descriptions to come.
The UT Austin team is:
- Aaron Chevalier
- Dan Blick
- Eric Davidson
- Jeff Tabor
- Laura Lavery
- Matt Levy
- Rachel Haurwitz
- [http://www.mine-control.com/ Zack Booth Simpson]
Advisors:
- [http://ellingtonlab.org/ Andy Ellington]
- [http://polaris.icmb.utexas.edu/home.html/ Edward Marcotte]
Light Wires
We are looking to build circuits which demonstrate the unique regulatory capabilites of the light dependent system developed by the UT/UCSF team last year. One such instantiation is the original [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5407 Edge Detector] (see below) from last year which we are finally finishing.
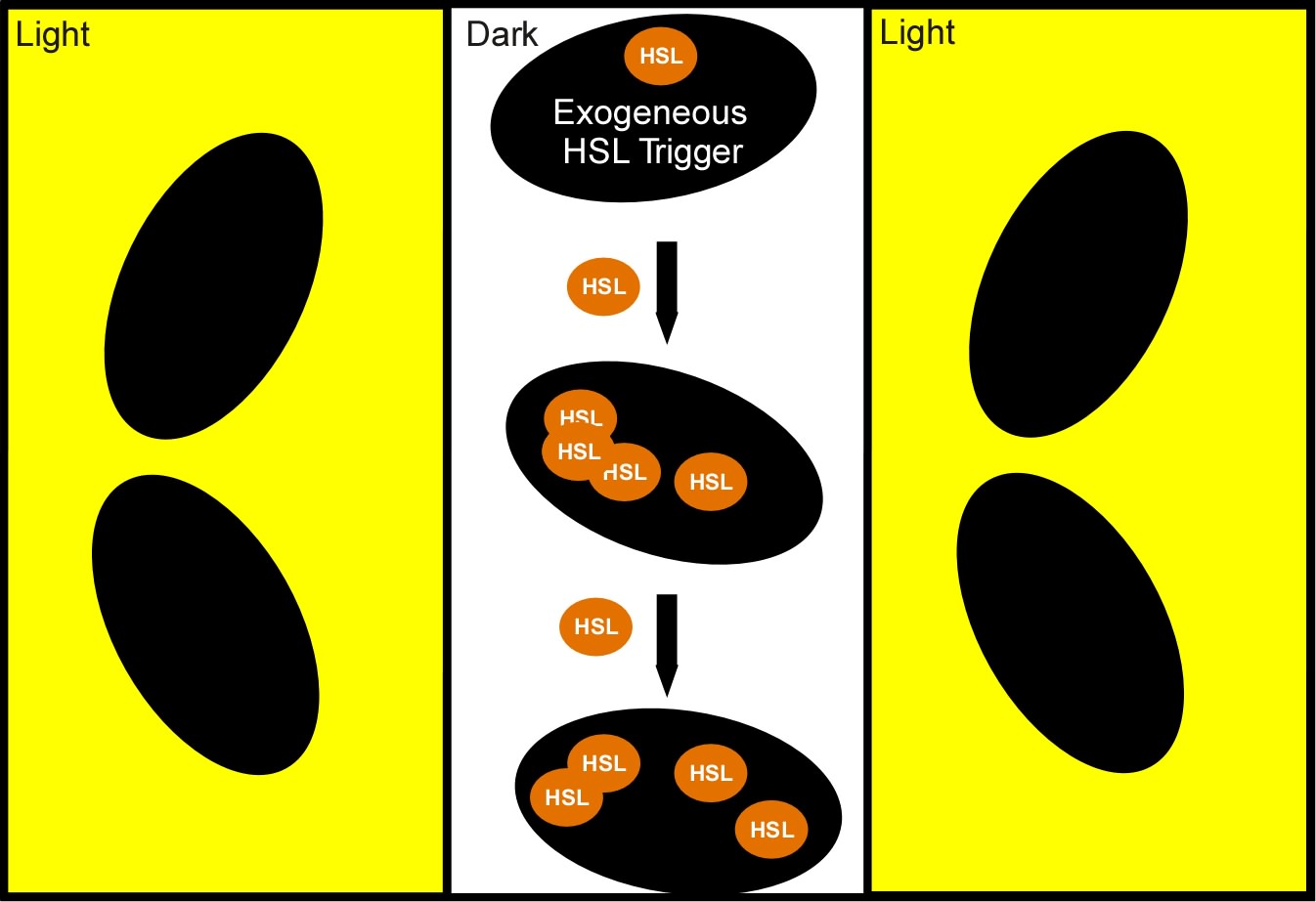
The idea which came out on top of this year's heap was to transform a genetically identical lawn of E.coli into a biochemical circuit board, using light to dictate the conductive potential of each ~1x3 micron segment of surface area. All cells on the lawn will carry two biochemical signal amplifiers: a 3OC6HSL amplifier [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5414 I15030], and a pseudomonas autoinducer (AI-1) amplifier [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5598 J13030]. These signal amplifiers will be inactive in certain locations of the plate and active in others. We will dictate which E.coli on the plate have active circuits with light, by using [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5597 J13100] to strongly repress these two signal amplifiers in the dark.
We will use light to draw intersecting wires on the E.coli canvas, and will activate biochemical signal propagation down each wire with a separate quorum sensing signal (see figure). The exogenous addition of a signal, say 3OC6HSL will activate high level production of that compound in the cells into which it diffuses. The newly produced 3OC6HSL will diffuse out of the biochemically active cells in all directions. Those cells which neighbor the activated cells AND which are in the light will similarly be triggered to actively produce the 3OC6HSL compound. This process will continue down the region of the agar plate, moving from cell to cell as far as the light allows.
We will propagate orthogonal biochemical currents down separate wires (figure), and will genetically encode different logical operations which will be relevant at the intersection of the two wires. The simplest logical operation will be an AND gate. If two orthogonal signals succesfully propagate down the plate and intersect at the end, two complementary halves of a reporter gene (LacZ) will be expressed and become functional, signaling an output of 1 from the AND gate.