4-State Device
From 2006.igem.org
Back to the ETH Zurich main page.
Contents |
Purpose
The 4-State Device (4-SD) uses two inputs to sequentially switch through its four states. The four states are used to keep track of changes of the inputs. One state is connected to the output.
Design
Suitable Strategies
For realizing the design we need one repressor for each state. For this several alternative can be considered: RNA interference to prevent translation of mRNAs, Using existing promotors as repressors, Designing new repressors using Zinc Finger domain.
Selected System
A RNA interference approach was abandoned since there are several problems could not be solved. If were to use known repressor such as tetR, we would have to modify the promoter regions of the lambda-system, Pr and Prm. Although a valid approach it is dropped, due to increased workload, cost and lack of modularity
We chose the approach use design new repressors using zinc finger domains. Furthermore, we put the binding sites for the repressors directly after start of transcription and before the ribosome binding site to prevent RNA polymerase to transcribe the gene. This would allow us to put any promotor in from of the binding site and allowing a very high degree of modularity.
There is possibility to use several 4-SD connected to each order in order to count n^2. but for every new device four new repressors is needed. With zinc finger domains the construction of any binding site is possible and we would not run out of repressor.
A Zinc Finger (ZF) [1] is a protein domain [2] that binds to three base pairs of double stranded DNA. A Zinc Finger Protein (ZFP) consists of one or several zinc finger domains. Many protein-DNA interaction for ZF domains and triplet of base pairs have been described, therefore making it possible to to construct artificial transcription factors by combining ZF domains in a modular fashion.
Actual Implementation
The central part of the counter is the 4SD. When input changes the next state is reached and the repressor is expressed. Every Repressor represses all other states except for the next. For example R1 represses R3 and R4, but not R2. This forces a progression of states R1 to R4 during the cycles of the input.
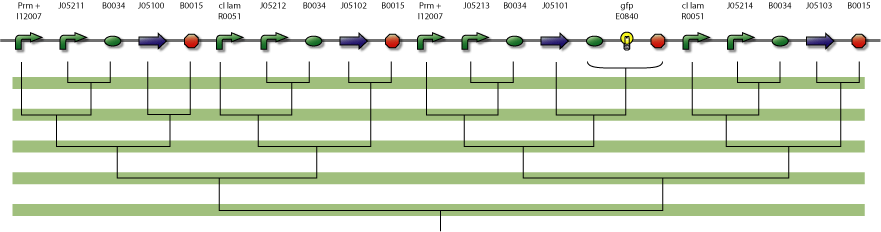
In the figure below is a parts-view of the device with PoPS interfaces (i/o).
Robins insertion start
Basic Functionality
The 4-State Device uses two inputs to sequentially switch through its four states. Note that this behaviour can be observed abstractly in the concept section.
To achieve such behaviour in our system, we use four interconnected gates. In the following, we call them NOR gates. Each of our NOR gates has three inputs. All of them have to be low for the output to be high.
Electrical NOR gate
A NOR gate is a very common part in electrical engineering.
______________
rs1---->¦ ¦
rs2---->¦ NOR gate ¦---->
rs3---->¦______________¦
Biological NOR gate
Biologically, a NOR gate can be implemented through a promoter with high basal activity that is repressed by three effectors. If there is at least one of the three repressors, transcription is inhibited. After each of the four NOR gates, there is a coding region for a protein. These four different proteins are called R1 to R4 in the following. (Note that such a biological NOR gate has no POPs interfaces)
biological picture of NOR gate
4-State Device circuit diagram
circuit diagram
4-State Device parts diagram
parts diagram
By design of the Event Processing Device, input 1 and input 2 have opposite activity, meaning that either R1/R3 or R2/R4 is active. Furthermore, since R1/R3 (respectively R2/R4) are repressing each other, only one of the two is active. Therefore, in a stable situation, only 1 of the 4 repressor proteins is expressed.
Let us assume that R1 is being expressed. Input 2 must then be low, and therefore input 1 high. This situation is stable and remains until there is a change in the inputs. Now, if input 1 decreases, and input 2 increases, the expression of R1 will come to a halt. Since input 1 is now low, either R2 or R4 will be expressed. At this stage, R1 is still present in relatively high concentration and by repressing R4, it tips the balance in favor of R2, leading to a new stable state in which only R2 is expressed.
Note that electrical engineers call such a device a "J-K flip flop". It can also be seen as a combination of two toggle switches (Atkinson03), each being able to store one bit.
Robins insertion end
In the design of the repressor protein we use the following domains:
Zinc Finger (ZF) DNA-binding domain
NTD The N-terminal polymerase domain from NusA. This protein has a RNA polymerase halting properties.
ERD
ERF repressor domain. a repressor domain in eukaryotes.
LZ
Leucine zipper. For dimerization.
If it turns out that repression has been used in all the literature we can find, then we would be better off to play safe and include them:
- Beerli PNAS 1998: they fused ZF to KRAB repressor and it has a stronger effect compare to ZF alone (see p14632 graph A)
- Beerli Nat Biotech review feb2002 : if you read the complete paragraph on gene repression (p 132), it gives strong evidence that we should fuse the ZF to a repressor (apparently at the N-Term of the ZF)
It is stated that polymerase blockade through ZF only is not very efficient. We can not be sure that the 45 amino acid long KRAB domain will work, if it doesn't exist in prokaryotes. KRAB domains have a very specific interaction with a co-repressor molecule. It might be better to find something already known to work in bacteria or skip the repressor approach altogether.
table of parts.
We use several different designs of ZFPs.
- Three ZF domains to recognize nine base pairs.
- Two ZF domains and a leucine zipper domain. The leucine zipper will be cause the protein to home-dimerize and hence it will recognize 12 base pairs.
- Six ZF domains to recognize twelve base pairs.
- Three ZF domains fused to negative transcription factors. (e.g. NTD and ERD)
| Nickname | Length | SacI | KpnI | NheI | EcoRI | XbaI | SpeI | PstI |
|---|---|---|---|---|---|---|---|---|
| BBa_J05215+BBa_J05108 | $874.35 | 1 | 91 | 598 | 7, 97 | 22, 112 | 71, 578 | 85, 592 |
| BBa_J05216+BBa_J05109 | $843.9 | 1 | 91 | 577 | 7, 97 | 22, 112 | 71, 557 | 85, 571 |
| BBa_J05217+BBa_J05110 | $883.05 | 1 | 91 | 604 | 7, 97 | 22, 112 | 71, 584 | 85, 598 |
| BBa_J05218+BBa_J05111 | $870 | 1 | 91 | 595 | 7, 97 | 22, 112 | 71, 575 | 85, 589 |
| BBa_J05221+BBa_J05101 | $722.1 | 1 | 112 | 493 | 7, 118 | 22, 133 | 92, 473 | 106, 487 |
| BBa_J05114 | $1238.3 | 1 | 849 | - | 7 | 22 | 829 | 843 |
| BBa_J05112 | $759.8 | 1 | 519 | - | 7 | 22 | 499 | 513 |
| BBa_J05222+BBa_J05115 | $980.2 | 1 | 87 | 671 | 7, 93 | 22, 108 | 67, 651 | 81, 665 |
describe the trick we use to reduce cost, that is, putting small parts together with big parts.
binding of ZFP. describe choice of binding sites. Describe the function of RNA pol II. emphasize: modularity! Another important aspect is the overall goal of keeping the design modular - one of the most important aspects of the this contest.
We have two alternatives for placing the binding sites.
1. Binding sites in the promoter. This would prevent the polymerase from binding to the promotor. Although this might be most likely to work, we have chosen to not pursue this alternative, while we are quite intrigued by the possibility of a roadblock. The operator regions (i.e. the "roadblocks" that will prevent of the RNApolymerase to transcribe the gene) form a BioBrick that should be inserted between the promoter region and the ribosome binding site in order to keep the design modular.
2. Binding sites directly after the start of transcription and before the ribosome binding site. This alternative is attractive, since it would allow for a high degree of modularity. In theory the ZFP would act as an extra "roadblock-operator" and any promotor could be used in front of the protein.
The ZFP roadblock operator regions (from now on refered to as: operators) consists of binding motifs for two repressors. The two bindning motifs are usually spaced with 5 base pairs (gcgcg). Some data of the binding strength were available and we have chosen operators with the affinity estimated to K_d = 3-40 nM.
| Part | Binding Site Seq | # exact matches | Hamming Dist 1 | Hamming Dist 2 | Hamming Dist 3 |
|---|---|---|---|---|---|
| BBa_J05100 | ggaggggac | 1 | 132 | 3125 | 32788 |
| BBa_J05102 | ggaggcggg | 5 | 396 | 6293 | 47897 |
| BBa_J05101 | gggggcgag | 8 | 303 | 4955 | 41302 |
| BBa_J05103 | ggggccgga | 23 | 433 | 5606 | 45492 |
| BBa_J05108 | gtcccctccggaggggac | 0 | 0 | 0 | 0 |
| BBa_J05109 | ctcgcccccgggggcgag | 0 | 0 | 0 | 2 |
| BBa_J05110 | cccgcctccggaggcggg | 0 | 0 | 0 | 3 |
| BBa_J05111 | tccggccccggggccgga | 0 | 0 | 0 | 3 |
| BBa_J05115 | gggggcgaggggggcgag | 0 | 0 | 0 | 1 |
Kd values of interactions. put in table above.
Degradation...?
Tests: design all tests. To test whether our assumptions about using multiple zinc finger proteins (ZFP) as repressors (i.e. roadblocks) will actually work, we will build a tester/debugging device in parallel with the counter.
| Repressor | Operator | Comments |
|---|---|---|
| J05100 | J05212 | ... |
| J05100 | J05212 | ... |
4SD system
The 4SD system consists of 4 proteins with 4 operator regions. It has an interface boundary with input module (Pr and Prm). Repressor R3 is connected to a reporter to be able to count to modulo two.
assembly diagram of device.