Abstraction hierarchy and PoPS
From 2006.igem.org
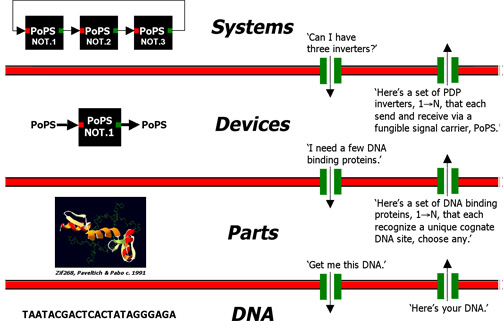
Abstraction Hierarchy
The purpose of an abstraction hierarchy is to hide information and manage complexity. To be useful, individuals must be able to work independently at each level of the hierarchy. In biology, for example, parts-level researchers might need to know what sorts of parts device-level researchers would like to use, how different types of parts actually work (e.g., atomic interactions between an amino acid and the major groove of DNA), and how to order a piece of DNA. But, parts-level researchers do not need to know anything aboutDNA synthesis chemistry, how short pieces of DNA are assembled into longer contiguous DNA fragments, or how a ring oscillator works, et cetera.
An abstraction hierarchy can be used with other technologies (e.g., 'standard part families') to quickly design and specify the DNA sequence encoding many integrated genetic systems. For example, a ring oscillator system can be built from three inverter devices. Each inverter device can in turn be built from four parts, and each part can be encoded by a pre-specified sequence of DNA. Other systems can be quickly specified as different combinations of devices, and so on.
Figure 1: Abstraction barriers (red) block all exchane of information between abstraction levels. Interfaces (green) enable the limited and principled exchange of information between levels. Example exchanges are given in quotes.
One detail of the abstraction hierarchy depicted in Figure 1 that's worth elaborating is the definition of input and output signals for gene-expression based devices. A device is a combination of parts that performs some useful function. For example, one type of device is an inverter. An inverter takes an input signal and produces the opposite output signal (e.g., HIGH input produces LOW output and vice versa; an inverter functions like a Boolean NOT).
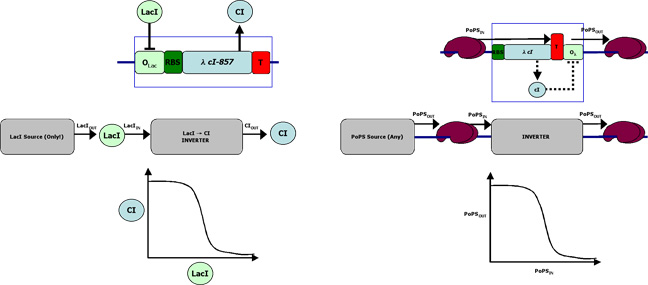
A genetic inverter can be assembled from four parts [Figure 2]. Classically, a genetic inverter receives as input the concentration of repressor A and, via gene expression, sends as output the concentration of repressor B [Figure 2, left]. In this model, the signal carriers are the proteins A and B, and the signal levels are the concentrations of proteins A and B. One immediate problem with such a device is that the input and output signal carriers are different. Such an inverter can only be connected to an upstream device that sends protein A and a downstream device that receives protein B. A full collection of such inverters would require all combinaations of protein A and protein B, the square of the number of proteins.
Polymerase Per Second - PoPS
We can solve this problem by reorganizing the parts that make up our inverter [Figure 2, right]. Now, the input signal to the inverter is carried by polymerase per second, or PoPS. PoPS is the flow of RNA polymerase molecules along DNA (i.e., 'current' for gene expression). The PoPS level is set by the amount of RNA polymerase molecules that trundle past a specific position on DNA each second. Importantly, the output signal from the inverter is also carried by PoPS. The inversion function of the device is still executed by a repressor protein acting on its cognate site at the operator DNA, but all details specific to this interaction are internal to the device. As a result, an inverter only involves one protein and a complete set requires only one inverter per protein.
Figure 2:
(Left) A 'classical' genetically encoded inverter that takes as input the concentration of a repressor protein, LacI. In the presence of LacI, expression of the downstream cI gene is inhibited. In the absence of the LacI, expression of the downstream cI occurs via transcription initiating at OLac, producing a high CI output signal.
(Right) A PoPS-based inverter. When the input PoPS-level is high, CI is produced. CI acts at OLambda to keep the output PoPS-level low. The molecule-specific details of a PoPS-based inverter are internal to the device and can be hidden; PoPS-based devices can thus be used in combination with (i.e., connected to) and other PoPS-based devices.