Assembly
From 2006.igem.org
Contents |
Parts to Make:
HixL: TTCTGGAAAA CCAAGGTTTT TGATAA
HixR: TTATCAAAAA CCTTCCAAAA GGAAAA
HixC: TTATCAAAAA CCATGGTTTT TGATAA
Backwards Parts
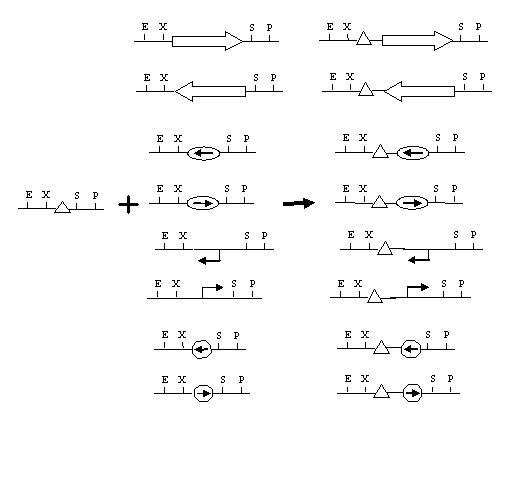
Because we wish to start with elements that are reversed (upside down pancakes), we need to think about how to get backwards parts. That is, parts that have the Prefix, then reversed element (promoter, term, CDS, RBS), then Suffix. Several possible ways:
- Some parts are backwards in the registry - terminators, for eg.
- Parts that are not long could be synthesized backwards.
- If one pancake can be flipped, reversed parts can be made. This may actually be a good contribution, since it could be applied to reverse any part between two hix sites.
- Can cut out a part with NotI, then religate in both orientations.
Examples
BBa_B0022 Terminator (Reverse B0012) Length: 84 Base Pairs Including BioBrick Ends
Bingo! BBa_P1004 is ampR resistance cassette in reverse orientation Length: 984 Base Pairs Including BioBrick Ends
Bba_B0034 is a RBS with the sequence:
gaattcgcggccgcttctagag aaagaggagaaa tactagtagcggccgctgcag
It could be redesigned with the RBS reversed:
gaattcgcggccgcttctagag tttctcctcttt tactagtagcggccgctgcag
Steps
1. Design/Order DNA – Hix L, R, & C
- Want to do a Left to Right BioB addition: Promoter + RBS w/AmpR -->Hix site-->
Term-->Hix(C)site-->TetR + Term
2. On receiving – restrict/ligate BioBricks into plasmid w/resistance gene on one side
a. Promoter, RBS BioB
b. TetR BioB (Backwards)
c. Term BioB
3. Transform E. coli
E = G/AATC
X = T/CTAGA
P = CGAT/CG
S = A/CTAGT
hix BioBricks
E - N - X - hix - S - N - P
Does there have to be a T between NotI and XbaI and an A between SpeI and NotI? All the parts we have looked at so far have this.
HixL
GAATTC GCGGCCGC T TCTAGA TTCTTGAAAACCAAGGTTTTTGATAA ACTAGT A GCGGCCGC CTGCAG
HixR
GAATTC GCGGCCGC T TCTAGA TTATCAAAAACCTTCCAAAAGGAAAA ACTAGT A GCGGCCGC CTGCAG
HixC
GAATTC GCGGCCGC T TCTAGA TTATCAAAAACCATGGTTTTTGATAA ACTAGT A GCGGCCGC CTGCAG
Oligos to order
HixL_top
AATTCGCGGCCGCTTCTAGATTCTTGAAAACCAAGGTTTTTGATAAACTAGTAGCGGCCGCCTGCA
HixL_bottom
GGCGGCCGCTACTAGTTTATCAAAAACCTTGGTTTTCAAGAATCTAGAAGCGGCCGCG
HixR_top
AATTCGCGGCCGCTTCTAGATTATCAAAAACCTTCCAAAAGGAAAAACTAGTAGCGGCCGCCTGCA
HixR_bottom
GGCGGCCGCTACTAGTTTTTCCTTTTGGAAGGTTTTTGATAATCTAGAAGCGGCCGCG
HixC_top
AATTCGCGGCCGCTTCTAGATTATCAAAAACCATGGTTTTTGATAAACTAGTAGCGGCCGCCTGCA
HixC_bottom
GGCGGCCGCTACTAGTTTATCAAAAACCATGGTTTTTGATAATCTAGAAGCGGCCGCG
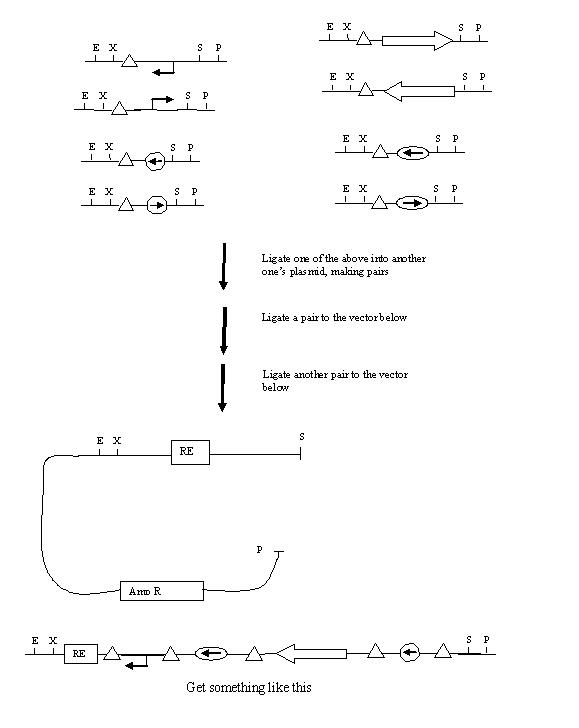
Building
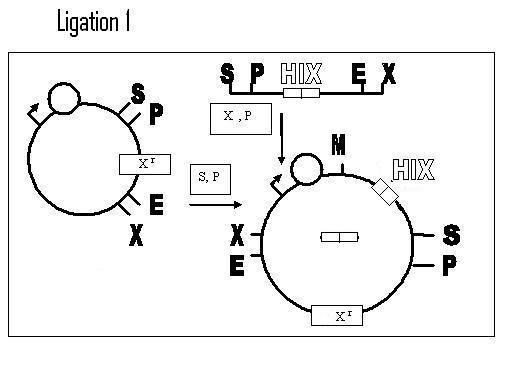
Biobricking in one hix site:
Example of a construct for one flip (terminator, although CDS would be better):
Biobricking hix sites onto parts:
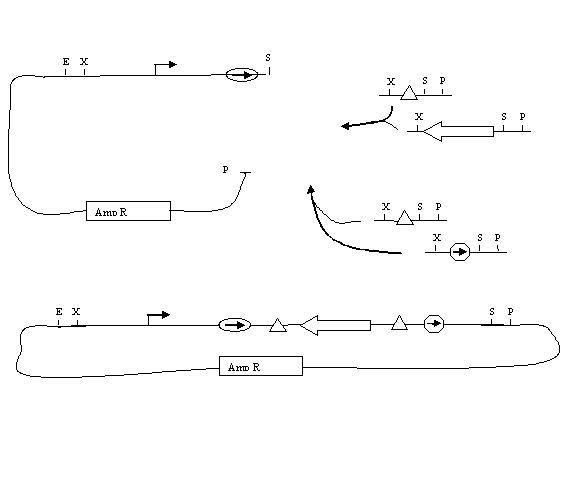
Assembly of One Pancake Test Construct:
Assembly of Two Pancake Test Construct would be next. Probably want two genes.
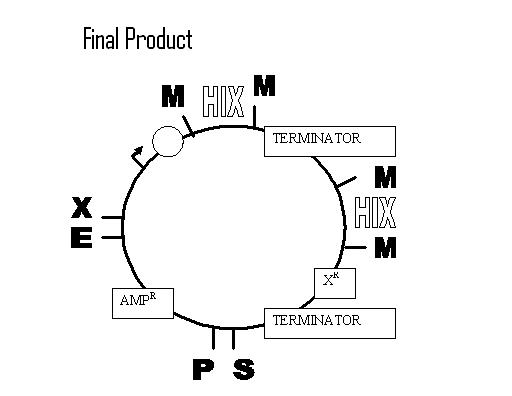
Assembly of Multiple Pancake Test Constructs:
First Transformations:
>1. Terminator T1 Bba_B0010 DNA-2 3P<
2. Promoter Pbad BBa_I13453
>3. Promoter w/RBS (TetR Repressed) BBa_J13002 DNA-2 3F <
4. Reverse Amp^r BBa_P1004 (Oops, planning)
5. Reverse CmR BBa_P1009 789 bp (Oops, planning)
>6. Double Forward Terminator BBa_B0015 DNA-1 1I <
>7. Tet^r Bba_P1001 DNA-2 23F <
Questions:
Will the TetR repressed promoter in BBa_J13002 be always on in JM109 cells?
Do we want to be able to turn on a Start Promoter(i.e. LacP using IPTG) on/off so recombination occurs 1st; and then use a different inducible promoter to turn on transcription?
Steps:
Step 1: Transform E. coli w/:
TetR (BBa_P1001 Plate DNA-2, Spot 3F)
Double Forward (Term) (BBa_B0015 Plate DNA-1, Spot 1I)
pBad (BBa_I0500, Plate DNA-2, Spot 9I)
pLac (BBa_R0011, Plate DNA-1, Spot 7M)
pSB1A3 (BBa_I13453, Plate DNA-2, Spot 13D
Step 2: Plasmid Preps.
Step 3: Use NotI to reverse TetR Gene
Step 4: Clone Hix DNA
Step 5: Assemble BioBricks