DNA Pancake
From 2006.igem.org
Defining a Biological DNA Pancake
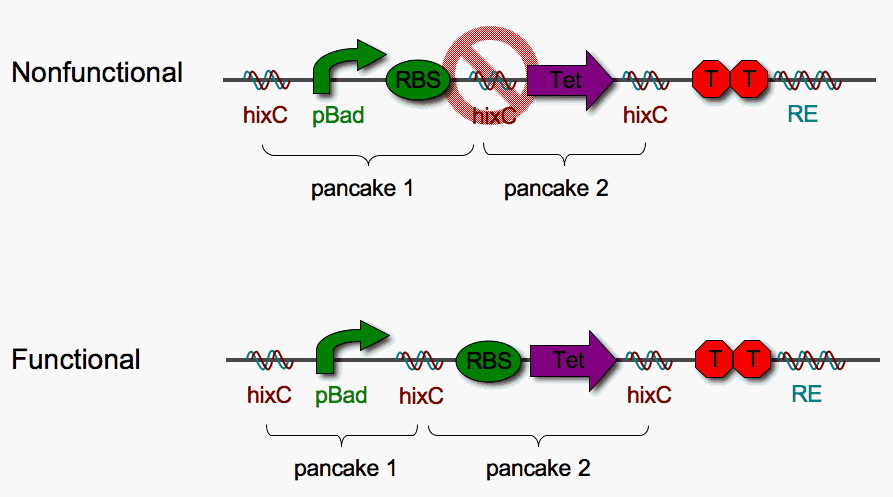
We chose basic BioBrick parts to create flippable DNA pancakes. Our DNA pancake constructs consist of the following units:
- pBad promoter : induced by arabinose C. We chose an inducible promoter to prevent constitutive transcription from interrupting Hin-mediated flipping
- RBS : The Ribosomal Binding Site is required for transcription of a coding sequence
- Tet : The tetracylcine resistance coding region
The Recombinational Enhancer (RE) facilitates Hin-mediated flipping. The double terminator was included to block read-through into the RE.
The bacteria will survive in the presence of tetracycline only when pBad, RBS and Tet are assembled in order and in the proper orientation. If they are scrambled in any way, the cells will die. Thus, if we start with bacteria carrying an artificially scrambled stack and allow flipping by adding Hin, we can simply plate these bacteria on tet agar and look for survivors. These bacteria have successfully unscrambled the stack.
Placing a BioBrick in between two Hix sites might disrupt the function of that BioBrick. To test this, we assembled some protoype "solved" stacks and looked for expression of Tet resistance. As shown above, the [pBad-RBS] [Tet] pancake stack is nonfunctional (not tetracycline resistant) while the [pBad] [RBS-Tet] stack is functional (tetracycline resistant). Therfore [pBad] and [RBS-Tet] define a functional biological 2-pancake stack. The remainder of our project is based upon these two units.
A two-pancake problem isn't very challenging, but it is a good starting point to demonstrate proof-of-concept. Once our system is established with this two-pancake stack, we can simply add more pancakes (Hix-flanked BioBricks) to asseble larger stacks with phenotypically unique solutions.