Event Processing Device
From 2006.igem.org
Contents |
Organisational
Group Members
- Christophe, Dominic (coordinator), Giorgia, Herve, Kristian, Zlatko
Group Meeting History
log 2005-08-17: Wednesday, 13:00 @ polyterrasse: Discussion of module, next steps, task distrib. log 2005-08-22: Monday, 15:00 @ polyterrasse: Discussion of biol. solutions.
Input-module Development
Module Description
In a nutshell, this module has 2 system boundaries, both of which are characterized by PoPS. Its purpose is to take a single input PoPS and output 2 different PoPS. One of the outputs should be high and the other low when S is high and vice versa when S is low.
PoPS_outn
^
¦
high ¦ PoPS_out2 _______ _________ PoPS_out1
¦ \ /
¦ \ /
¦ \ /
¦ \ /
¦ \ /
¦ X
¦ / \
¦ / \
¦ / \
¦ / \
low ¦ PoPS_out1 _______/ \_________ PoPS_out2
¦--------------------------------------------------> PoPS_in, t
0 1
Input-module Schematic
Below a preliminary parts-view of the module, i.e. encapsulation of biological specific implementations into a functional box with general PoPS interfaces.
-----------------------
¦ ¦
¦ ,--- a ---. ¦
¦ / ¦ ¦
-------- ¦------- act V ¦
¦ ¦ ¦¦ ¦ -- P_a -- PoPS_out1 --> to R_1,R_3
I -> ¦ P_in ¦-- PoPS_in -->¦¦ Reg ¦ ¦
¦ ¦ ¦¦ ¦ -- P_r -- PoPS_out2 --> to R_2,R_4
-------- ¦------- rep --- ¦
¦ \ ¦ ¦
¦ `--- r ---' ¦
¦ ¦
----------------------¦
legend: I: input signal that will bind to the promotor P_in, e.g. heat shock dependent etc. P_in: promotor that allows the signal of choice I to bind. PoPS_in: polymerase per second dependent on promotor and concentration of I Reg: regulation genes. there are different solutions possible, see below. r: repressing signal. highly dependent on Reg, P_r and of course speed and binding considerations a: activating signal. highly dependent on Reg, P_a and of course speed and binding considerations rep: repression, indicated by horizontal bar act: activation, indicated by arrow P_r: promotor region to be repressed and/or "roadblock" region. constitutively active P_a: promotor region to be activated and/or "roadblock" region. not constitutively active PoPS_out1: polymerase per second dependent on P_r and r. PoPS_out2: polymerase per second dependent on P_a and a. R_n: toggle switch gene(s)
Possible Implementations
Check the following page for lists of transcriptional regulators in E.Coli: [1] [2]
Two Regulators (a,r)
In the strategies below, the input PoPS regulates two proteins, an activator and a repressor. That has the advantage of being very simple, but requires that both proteins have roughly similar production and degradation speed.
Basic (Giorgia)
We will keep this strategy as our "last shore", since it is quite banal. Basically, one activator and one repressor genes will be under control of the same promoter, thus the encoded proteins will be produced in parallel. The two regulators will then act separately on their respective promoters. The tricky part of this strategy would be to find two regulators that activate/repress their promoters with similar "strength".
Antisense (Giorgia)
- Background:
In antisense strategy, repression of protein production takes place at the translational level. An antisense gene has a sequence exactly "opposite" (reversed, complementary) to the sequence of the gene that should be repressed. Transcription of both the antisense gene and the sense gene occurs, so that sense mRNA and antisense mRNA are produced. The "antisense" mRNA molecule binds tightly to its mirror image, thus preventing translation.Activation is achieved by means of an activator/promoter set, e.g. the AraC/pBAD.
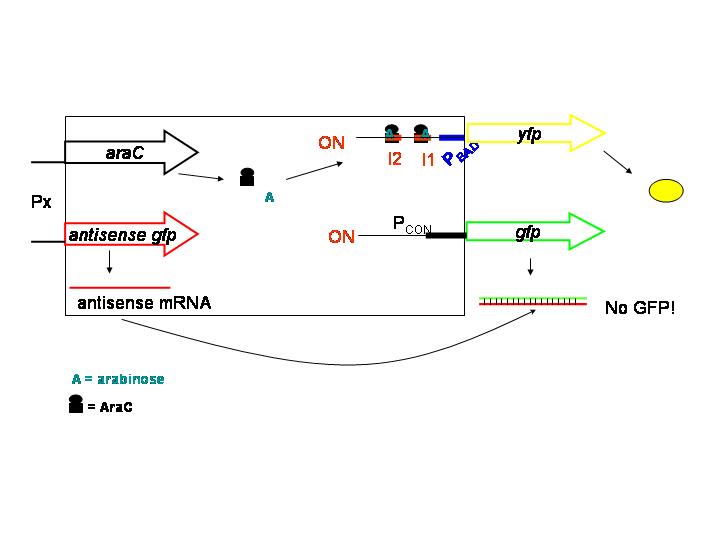
- Problems:
- How efficient is repression of translation? In bacteria, ribosomes bind while the DNA is being transcribed into mRNA.
- Coordination of repression and activation could be a problem, due to a delay in repression.
One Regulator (ar)
In the implementations below, the input PoPs regulates the production of a sole protein that somehow acts as activator and repressor. With only one regulator, production and degradation are perfectly uniform, which is good, but in some cases, it might be harder to obtain a symmetrical activity, which is bad.
"lambda" (Giorgia)
- Background:
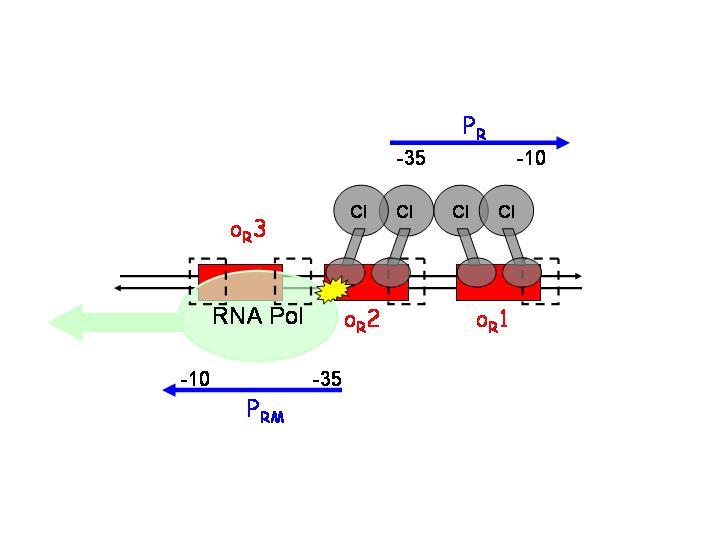
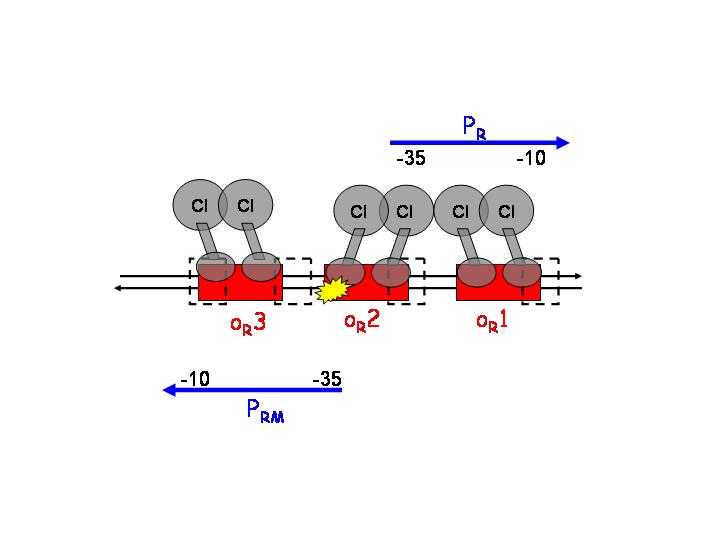
This strategy is based on the pR and pRM promoters of lambda phage. The input PoPs regulates the production of CI, which will act as an activator of pRM and as a repressor of pR.
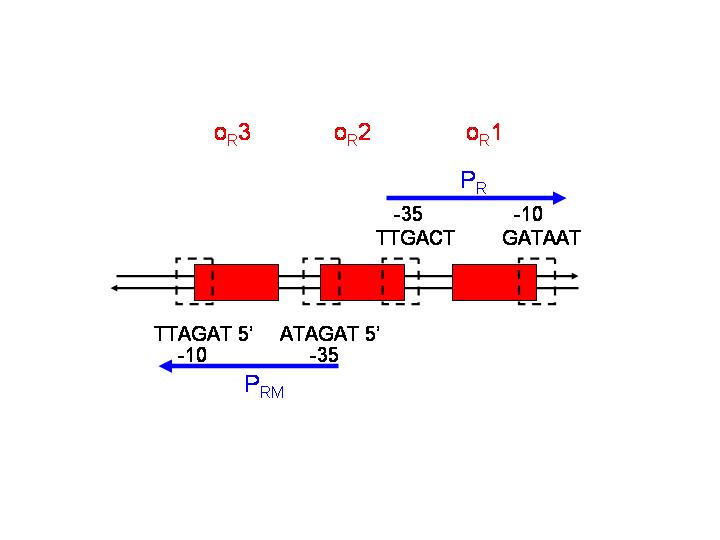
pR is a constitutive promoter, which is repressed by cooperative binding of two CI dimers onto the operator sequences OR1 (highest affinity) and OR2. On the other hand, pRM is activated by CI binding to OR1 and OR2.
After CI concentration reaches a threshold, binding to OR3 (lowest affinity) and inactivation of pRM occurs.
- Pros:
- Since the two promoters are regulated by the same protein-operator interactions, repression and activation should be symmetrical.
- pR, pRM and CI are available from the registry.
- Open questions:
- Should OR3 be removed from the pRM sequence? Or would the late-repression of pRM not jeopardise the achievement of our desired behaviour?
- Which sequence could be adequate to replace OR3 (spacing between -35 and -10 of pRM has to be maintained)? Would this replacement influence the binding affinity of CI to OR1?
- Basal activity of pRM?
AraC (Giorgia)
This option was dumped after further literature reading.
"Lunatic Activator" (Christophe)
The "Lunatic Activator" is an activator because it is required for the transcription of gene 1 and 3, but is lunatic because it can also acts as a roadblock on genes 2 and 4. Send me an email if you know such protein.
Zinc Finger YY1 (Hervé)
Secondary Metabolism (Christophe)
The idea here would be to use an existing regulator that both activates and represses genes required in the secondary metabolism (e.g. Amino-acid production). To minimize interference with E.Coli, it would be wise to try a system from a very different bacteria (e.g. B. Subtilis).