Ljubljana, Slovenia 2006/Methods
From 2006.igem.org
| (62 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <center>[[Image:Logo-si1 | + | <center>[[Image:Logo-si1.gif|center|120 px]] <br> |
[[Image:line-si4.jpg]] | [[Image:line-si4.jpg]] | ||
<table cellpadding="10"> | <table cellpadding="10"> | ||
| - | <tr><th>[[ | + | <tr><th>[[Ljubljana, Slovenia 2006|Home]]</th> |
| - | <th>[[Background and Signalling Pathway]]</th> | + | <th>[[Ljubljana, Slovenia 2006/Background and Signalling Pathway|Background and Signaling Pathway]]</th> |
| - | <th>[[Project]]</th> | + | <th>[[Ljubljana, Slovenia 2006/Project & Model|Project & Model]]</th> |
| - | <th>[[Results & Conclusions]]</th> | + | <th>[[Ljubljana, Slovenia 2006/Results & Conclusions|Results & Conclusions]]</th> |
| - | <th>[[Terms & References]]</th> | + | <th>[[Ljubljana, Slovenia 2006/Terms & References|Terms & References]]</th> |
| - | <th>[[Team members]]</th></tr></table> | + | <th>[[Ljubljana, Slovenia 2006/Team members|Team members]]</th></tr></table>[[Image:line-si3.jpg]] |
| - | [[Image:line-si3.jpg]] | + | |
<br> | <br> | ||
</center> | </center> | ||
| Line 16: | Line 15: | ||
<h2>Parts design</h2> | <h2>Parts design</h2> | ||
| - | <p>At first we had to design primers to replicate a desired DNA fragments. In primers we included restriction sites - on left site XbaI and on the right site SpeI, NcoI and PstI. We cloned that part into BioBrick plasmids with ccdB domain to get all restriction sites needed for BioBrick assembly. We had to design all parts de novo, since no parts like promoters, terminators, desired proteins for signaling pathway modification, degradation flags and reporters had been designed so far - neither to work in mammalian cells. List of desired constructs is shown below. For our use we designed a special vector ([http://partsregistry.org/Part:BBa_J52017 BBa_J52017]) with terminator to simplify constructs assembly. All our composite parts (promoter plus part) were then cloned in this vector.</p> | + | <p>At first we had to design primers to replicate a desired DNA fragments. In primers we included restriction sites - on left site ''XbaI'' and on the right site ''SpeI'', ''NcoI'' and ''PstI''. We cloned that part into BioBrick plasmids with ''ccdB'' domain to get all restriction sites needed for BioBrick assembly. We had to design all parts ''de novo'', since no parts like promoters, terminators, desired proteins for signaling pathway modification, degradation flags and reporters had been designed so far - neither to work in mammalian cells. List of desired constructs is shown below. For our use we designed a special vector ([http://partsregistry.org/Part:BBa_J52017 BBa_J52017]) with terminator to simplify constructs assembly. All our composite parts (promoter plus part) were then cloned in this vector.</p> |
| - | <p>We also needed fusion proteins e.g. | + | |
| + | <p>We also needed fusion proteins e.g. [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]]-rLuc-PEST ([http://partsregistry.org/Part:BBa_J52013 BBa_J52013]) - that is our dominant negative protein linked with reporter and degradation flag. This parts are designed like basic parts - not composite, although they are fusion proteins. Between proteins there is a six aminoacid long linker because only two aminoacid long linker formed during biobrick assembly could affect protein folding. We introduced a six amino acids long linker inbetween protein - reporter and reporter - degradation flag with primers using PCR Overlap Extension method. These parts were then combined with promoter ([[Ljubljana, Slovenia 2006/Terms & References|NF-κB]]) in BioBrick assembly technique. The part was then inserted into the vector with terminator and ready to use in human cells ([[Ljubljana, Slovenia 2006/Terms & References#Terms|HEK293]]).</p> | ||
| + | <br> | ||
<h2>Experiments on mammalian cell cultures</h2> | <h2>Experiments on mammalian cell cultures</h2> | ||
| - | <p>In September and October we were transfecting our constructs into human embrional kidney cells ([[ | + | <p>In September and October we were transfecting our constructs into human embrional kidney cells ([[Ljubljana, Slovenia 2006/Terms & References#Terms|HEK293]]). At the same time we were optimizing three detection systems mentioned below. At first we had to optimize the methods (read articles, test negative and positive controls) and learn how to work with human cells.</p> |
| - | <p>Cells were transfected not only with our constructs but also with TLRs which are not natively present on the surface of [[ | + | |
| - | + | <p>Cells were transfected not only with our constructs but also with TLRs which are not natively present on the surface of [[Ljubljana, Slovenia 2006/Terms & References#Terms|HEK293]] cells (only TLR3 and TLR6 are present).</p> | |
| - | + | ||
| + | <p>First we transfected all [[Ljubljana, Slovenia 2006/Terms & References#Terms|HEK293]] cells with our constructs and with two additional plasmids coding for [[Ljubljana, Slovenia 2006/Terms & References#Terms|TLR4]] and the accessory [[Ljubljana, Slovenia 2006/Terms & References#Terms|MD-2]] protein. Very soon, we found out that our plasmid preparations were contaminated with [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]] from ''E. coli'' (strain DH5α used for plasmid preparation), since there was no difference in the response between stimulated (with [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]]) and unstimulated cells. To overcome this problem we decided to focus on TLR5 receptor which reacts upon contact with bacterial [[Ljubljana, Slovenia 2006/Terms & References#Terms|flagellin]]. Signal transfer through TLR5 does not depend on the presence of [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]].</p> | ||
| + | <br> | ||
<h2>Detection systems</h2> | <h2>Detection systems</h2> | ||
| - | <p>In order to monitor the | + | <p>In order to monitor the activation of the signaling pathway and function of our Parts we considered using several detection systems. We expected the synthesis of dominant negative protein ([[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] or [[Ljubljana, Slovenia 2006/Terms & References#Terms|TRAF6]]), blocking of signaling pathway and consequently transcription termination of dominant negative protein. The constructs (parts) we prepared were designed for each detection system respectively.</p> |
<p>Requests for the optimal detection system were: | <p>Requests for the optimal detection system were: | ||
| Line 46: | Line 49: | ||
<p>Constucts used in individual detection system and short description of detection systems:</p> | <p>Constucts used in individual detection system and short description of detection systems:</p> | ||
| + | |||
| + | <br> | ||
<h4>Flow cytometry</h4> | <h4>Flow cytometry</h4> | ||
| - | Used composite parts: NFκB- | + | Used composite parts: NFκB+dnMyD88-link-rLuc ([http://partsregistry.org/Part:BBa_J52014 BBa_J52014]). |
| - | + | ||
| - | + | ||
| - | + | [[Image:Flow cytometer.jpg|right|thumb|200px|Figure 15: Ota working on the flow cytometer.]] | |
| - | <p> | + | <p>Flow cytometry is a sophisticated method, which gives very precise and highly valuable results. The main advantage of this method is that measurements are done on individual cells, so the results are not the average value of the population. In our project we could detect cellular activation based on the phosphorylation cascade activation and anti-phosphoprotein antibodies. In our experiments we used primary antibodies (Ab) from mouse, which specifically detect only phosphorylated form of [[Ljubljana, Slovenia 2006/Terms & References#Terms|ERK kinase]] (pERK). The secondary antibodies used were anti-mouse Abs labelled with a fluorescence marker phycoerithrin (PE), which we can detect on flow cytometer. Cells were transfected with TLR5 or TLR4/MD-2 and with or without [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]], which is under [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] promoter. On the second day the cells were stimulated with flagellin or [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]] (depends on the selected TLR receptor). According to the literature phosphorylation of [[Ljubljana, Slovenia 2006/Terms & References#Terms|ERK]] can already be detected after 15 min. The amount of p[[Ljubljana, Slovenia 2006/Terms & References#Terms|ERK kinase]] increases for additional 20 min and then it starts to decline. After 40 min no p[[Ljubljana, Slovenia 2006/Terms & References#Terms|ERK]] is detected.</p> |
| - | <h4> | + | <p>We used pulsed stimulation of cells. First we stimulated cells with 5 ng/ml of flagellin to activate the expression of [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]]. We repeated the stimulation with the second, stronger stimulus (50 ng/ml of flagellin) at different time intervals (2, 4, 6 and 8 hours after the first stimulation. 30 min after the second stimulus cells were fixed in 2% paraformaldehid, permeabilized with ice cold methanol and stained for p[[Ljubljana, Slovenia 2006/Terms & References#Terms|ERK]] and secondary Ab labelled with PE.</p> |
| + | |||
| + | <p>Expected results: The activation of the signaling pathway also activates the expression of [[Ljubljana, Slovenia 2006/Terms & References#Terms|dominant negative protein MyD88]], which should block the signaling, so we expect to detect decreased amount of p[[Ljubljana, Slovenia 2006/Terms & References#Terms|ERK kinase]] after second stimulation in comparison to cells without of inducible [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]].</p> | ||
| + | <br> | ||
| + | |||
| + | <h4>Detection of transcriptional activation by luminometry</h4> | ||
| - | Used composite parts: NFκB- | + | Used composite parts: NFκB+dnMyD88 ([http://partsregistry.org/Part:BBa_J52036 BBa_J52036]), NFκB+dnMyD88-link-rLuc ([http://partsregistry.org/Part:BBa_J52014 BBa_J52014]), NFκB+dnMyD88-link-rLuc-link-PEST191([http://partsregistry.org/Part:BBa_J52024 BBa_J52024]), NFκB+rLuc-link-PEST191([http://partsregistry.org/Part:BBa_J52023 BBa_J52023]), NFκB ([http://partsregistry.org/Part:BBa_J52010 BBa_J52010]), CMV-rLuc([http://partsregistry.org/Part:BBa_J52038 BBa_J52038]) and CMV+rLuc-link-PEST191([http://partsregistry.org/Part:BBa_J52039 BBa_J52039]). |
[[Image:luminometer.jpg|200px|right|thumb|Mithras luminometer]] | [[Image:luminometer.jpg|200px|right|thumb|Mithras luminometer]] | ||
| - | <p>Luminometry is a very used method to measure activation of signaling pathways. In many cases it replaced ELISA because it is much more sensitive. The basis of the method is the measurment of light, which is emited after substrate cleavage. We used two different enzymes | + | <p>Luminometry is a very used method to measure activation of signaling pathways. In many cases it replaced ELISA because it is much more sensitive. The basis of the method is the measurment of light, which is emited after substrate cleavage. We used two different enzymes; the firefly luciferase and the Renilla (sea pansy) luciferase can discriminate between their respective bioluminescent substrates and do not cross-activate, that is why we can measure the activity of both in one sample.</p> |
| - | <p> | + | <p>Dual luciferase assay</p> |
| + | <p>Dual luciferase assay includes two different luciferase reporter enzymes that are expressed simultaneously in each cell. Typically, the experimental reporter (in our case NFkB-fLuc) is correlated with the effect of specific experimental conditions, while the activity of the co-transfected "control" reporter gene (in our case CMV-rLuc) provides an internal control, which serves as the baseline response. Normalizing the experimental reporter gene to the activity of an internal control minimizes the variability caused by differences in cell viability and transfection efficiency. Thus, dual reporter assays allow more reliable interpretation of the experimental data by reducing extraneous influences.</p> | ||
| + | |||
| - | <p> | + | <p>Different experiments were conducted:</p> |
| - | <p> | + | <p>1. To be able to measure the activation of dnMyD88 expression after stimulation, we prepared dnMyD88 construct under [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] inducible promoter in the fusion with rLuc (construct NFkB+dnMyD88-link-rLuc). We transfected cells with TLR5 receptor plasmid and our biobrick construct. Cells were stimulated with [[Ljubljana, Slovenia 2006/Terms & References#Terms|flagellin]]. The expression of NFkB+dnMyD88-link-rLuc was measured in time intervals. We expect that amount of rLuc will increase in time only if cells are stimulated with a ligand.</p> |
| - | + | ||
| - | < | + | <p>2. CMV-rLuc-link-PEST191 biobrick was constructed to measure the kinetics of degradation of proteins in fusion with PEST. Cells were transfected with the construct. We found out that it takes four hours after the transfection to detect luciferase activity. To calculate half-life of rLuc and rLuc-PEST we transfected cells with either of those two constructs and incubated them to allow proteins to sythesize. New protein synthesis was inhibited by adding cycloheximide (inhibitor of protein synthesis in higher eucaryotes) and the activity of rLuc was measured in time intervals . Because the addition of cycloheximide stops the synthesis of new proteins there should be a decrease in rLuc activity due to protein degradation. A construct with [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]] should decrease more rapidly as rLuc alone.</p> |
| - | Used composite parts: NFκ | + | <p>3. Cells were transfected with TLR5, NFkB-fLuc, CMV-rLuc and +/- NFkB+dnMyD88. To have the amount of NF-kB binding sites equal in all cells we added the same amount of plasmid containing only [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] inducible promotor to cells without NFkB+dnMyD88. The next day after the transfection cells were first stimulated with 5 ng/ml of flagellin to activate the signaling pathway and thus dnMyD88 expression. After 6 hours (time, which was measured as an optimal time for detection of the expression of dnMyD88) cells were restimulated with 50 ng/ml of flagellin. Activation of fLuc was measured in time intervals (0, 2, 4, and 6 hours after the second stimulation). The data were normalized for transfection efficiency by measuring rLuc. We expect that there will be a lower activation after the second stimulation in cells, which express dnMyD88.</p> |
| - | <p>With ELISA detection system we are detecting free NF- | + | |
| + | <h4>ELISA for detection of transcription factors</h4> | ||
| + | |||
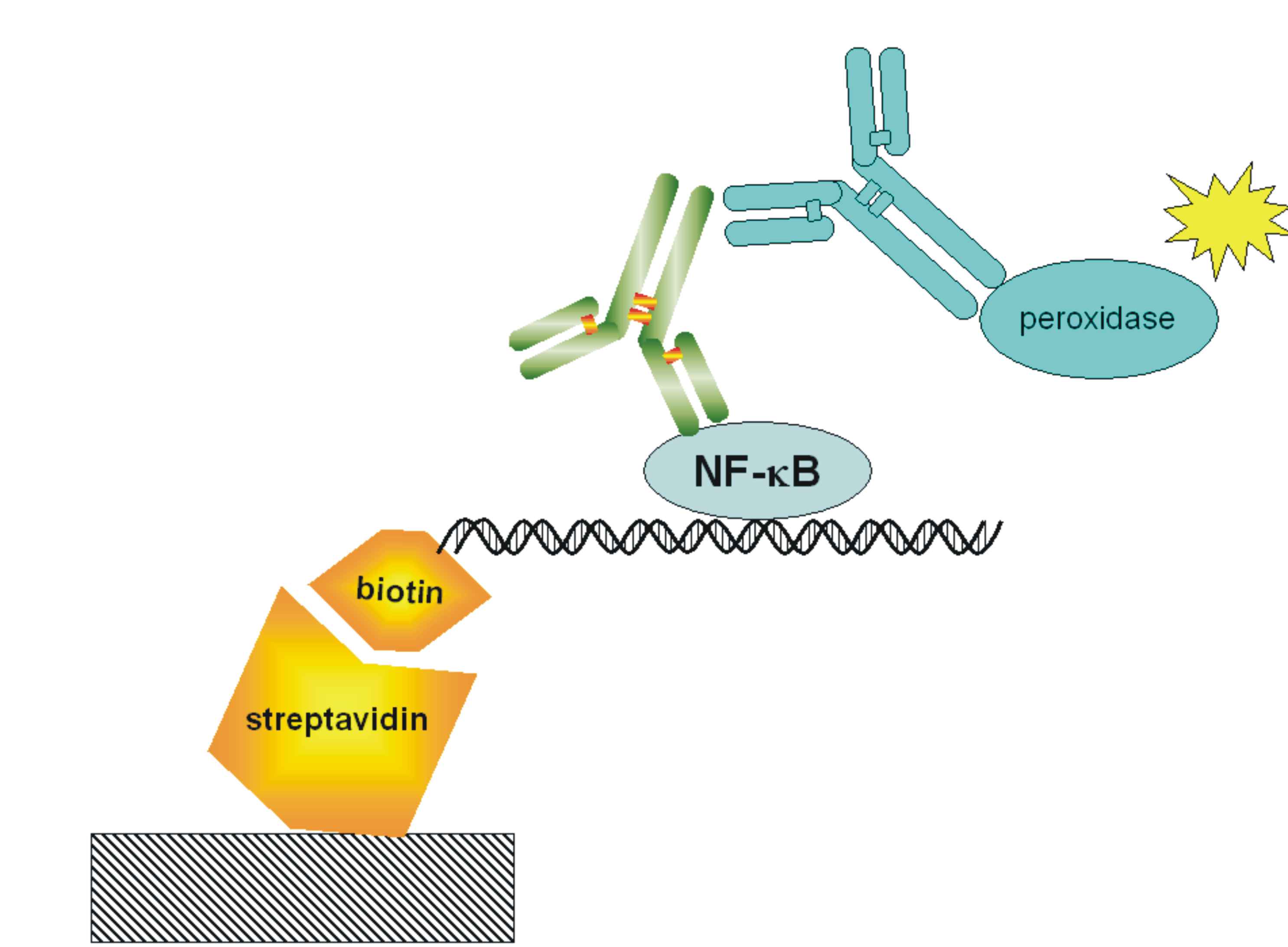
| + | [[Image:NF-kB-ELISA.jpg|200px|right|thumb|Figure 17: Schematic representation of the ELISA for detection of the transcriptionally-active NFκB.]] | ||
| + | |||
| + | Used composite parts: NFκB+dnMyD88 ([http://partsregistry.org/Part:BBa_J52036 BBa_J52036]) and NFκB+dnTRAF6-link–GFP ([http://partsregistry.org/Part:BBa_J52022 BBa_J52022]). | ||
| + | |||
| + | <p>With ELISA detection system we are detecting free [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] transcription factor, which it was already shown that it is a good measurment of pathway activation. Active form of this protein is present after the activation of signaling pathway when protein is released from a complex with IkB inhibitor. Free NF-kB migrates into the nucleus where it acts as transcription activator. We are using high bind 96-well plates coated with [http://en.wikipedia.org/wiki/Streptavidin streptavidin](we prepare it ourselves). The probe for free [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] is biotinilated double stranded [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] binding sequence. The free [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] protein from whole cell lysate is captured by a probe, which biotin group binds to streptavidin coated on a plate. We detect [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] with primary mouse antibodies and secondary peroxidase-conjugated rabbit anti-mouse antibodies. Peroxidase is an enzyme, which cleaves a substrate that emits light, which we measure on luminometer. </p> | ||
| + | <br> | ||
| + | |||
| + | <h4>Confocal microscopy</h4> | ||
| + | [[Image:celica2.gif|right|thumb|200px|Figure 18: Fluorescence of MyD88-GFP in transfected HEK293 cells.]] | ||
| + | <p>Microscope allows localization of signaling molecules within each individual cell, such as translocation of transcription factors, trafficking of proteins fused with reporter fluorescent proteins etc. Therefore this technique should be very usefull particularly for monitoring the expression of [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] inhibitory domain fused with fluorescent protein and the timecourse of its degradation. Therefore we designed Parts and several Devices, which included fusion with fluorescent protein:</p> | ||
| + | <br><br> | ||
| + | [http://partsregistry.org/Part:BBa_J52021 BBa_J52021] | ||
| + | <br>[http://partsregistry.org/Part:BBa_J52021 BBa_J52022] | ||
| + | <br>[http://partsregistry.org/Part:BBa_J52021 BBa_J52026] | ||
| + | <br>[http://partsregistry.org/Part:BBa_J52021 BBa_J52027] | ||
| + | <br>[http://partsregistry.org/Part:BBa_J52021 BBa_J52040] | ||
| + | <br>[http://partsregistry.org/Part:BBa_J52021 BBa_J52048]. | ||
| + | <br> | ||
<h1>Problems and Troubleshooting</h1> | <h1>Problems and Troubleshooting</h1> | ||
''Construction of Parts using bad oligos''<br> | ''Construction of Parts using bad oligos''<br> | ||
| - | We can only suggest to the other teams in the future to order oligos from a reliable and well established company (not necessarily the lowest price). When we repeatedly failed to produce PCR products it could have been the beginner's bad luck but when we determined a huge number of point mutations in the primer region even the mentors decided that we | + | <p>We can only suggest to the other teams in the future to order oligos from a reliable and well established company (not necessarily the onw with the lowest price). When we repeatedly failed to produce PCR products we thought it could have been the beginner's bad luck, but when we determined a huge number of point mutations in the primer region even the mentors decided that we better avoid this company in the future (although they did offer us the replacement of oligos for free). There'll be no next time for this company in our lab.</p> |
| + | |||
| + | ''Theory versus practise''<br> | ||
| + | <p>We spent a lot of time trying to find an answer to the following question: "Why didn't our transformants grow on the LB-Tc plates?". First we accused our our tetracycline, than to the proper concentration, but at the end we found out that one of the previous year's parts was incorrectly registered. Now we know the importance of checking everything twice... </p> | ||
| - | |||
| - | |||
| - | '' | + | ''Real-time luminescence''<br> |
| - | + | <p>We have attempted to monitor "real time" cell activation using luminescence and invested considerable amount of time and effort into this. We could not detectluminescence ''in vivo'' since coelentrazine (substrate of Renilla luciferaze) decays too quickly, while the luciferin could not penetrate the cells. We have also used the caged luciferin as well as its methy esther but in neither case did we suceed in detecting the luciferase in the living cells (several authors of published protocols were unavailable for comments :-( ).</p> | |
| - | |||
| - | |||
''Optimization of activation intervals''<br> | ''Optimization of activation intervals''<br> | ||
| - | Much od lab work (more than initially planned) was devoted to the | + | <p>Much od lab work (more than initially planned) was devoted to the design of experiments to monitor cell activation. We had to find out the optimal concentration of inducers, the timespan between the first and second activation which gave the best difference, lenght of the experiment in which difference between genetically engineered and wild-type cells would be sufficiently prominent etc. Methods from the literature could not simply be adopted into our experimental procedure.</p> |
<center> | <center> | ||
| Line 98: | Line 126: | ||
[[Image:line-si4.jpg]] | [[Image:line-si4.jpg]] | ||
<table cellpadding="10"> | <table cellpadding="10"> | ||
| - | <tr><th>[[ | + | <tr><th>[[Ljubljana, Slovenia 2006|Home]]</th> |
| - | <th>[[Background and Signalling Pathway]]</th> | + | <th>[[Ljubljana, Slovenia 2006/Background and Signalling Pathway|Background and Signaling Pathway]]</th> |
| - | <th>[[Project]]</th> | + | <th>[[Ljubljana, Slovenia 2006/Project & Model|Project & Model]]</th> |
| - | <th>[[Results & Conclusions]]</th> | + | <th>[[Ljubljana, Slovenia 2006/Results & Conclusions|Results & Conclusions]]</th> |
| - | <th>[[Terms & References]]</th> | + | <th>[[Ljubljana, Slovenia 2006/Terms & References|Terms & References]]</th> |
| - | <th>[[Team members]]</th></tr></table> | + | <th>[[Ljubljana, Slovenia 2006/Team members|Team members]]</th></tr></table> |
[[Image:line-si3.jpg]] | [[Image:line-si3.jpg]] | ||
<br> | <br> | ||
</center> | </center> | ||
Latest revision as of 10:10, 2 November 2006
| Home | Background and Signaling Pathway | Project & Model | Results & Conclusions | Terms & References | Team members |
|---|
Contents |
Parts design
At first we had to design primers to replicate a desired DNA fragments. In primers we included restriction sites - on left site XbaI and on the right site SpeI, NcoI and PstI. We cloned that part into BioBrick plasmids with ccdB domain to get all restriction sites needed for BioBrick assembly. We had to design all parts de novo, since no parts like promoters, terminators, desired proteins for signaling pathway modification, degradation flags and reporters had been designed so far - neither to work in mammalian cells. List of desired constructs is shown below. For our use we designed a special vector ([http://partsregistry.org/Part:BBa_J52017 BBa_J52017]) with terminator to simplify constructs assembly. All our composite parts (promoter plus part) were then cloned in this vector.
We also needed fusion proteins e.g. dnMyD88-rLuc-PEST ([http://partsregistry.org/Part:BBa_J52013 BBa_J52013]) - that is our dominant negative protein linked with reporter and degradation flag. This parts are designed like basic parts - not composite, although they are fusion proteins. Between proteins there is a six aminoacid long linker because only two aminoacid long linker formed during biobrick assembly could affect protein folding. We introduced a six amino acids long linker inbetween protein - reporter and reporter - degradation flag with primers using PCR Overlap Extension method. These parts were then combined with promoter (NF-κB) in BioBrick assembly technique. The part was then inserted into the vector with terminator and ready to use in human cells (HEK293).
Experiments on mammalian cell cultures
In September and October we were transfecting our constructs into human embrional kidney cells (HEK293). At the same time we were optimizing three detection systems mentioned below. At first we had to optimize the methods (read articles, test negative and positive controls) and learn how to work with human cells.
Cells were transfected not only with our constructs but also with TLRs which are not natively present on the surface of HEK293 cells (only TLR3 and TLR6 are present).
First we transfected all HEK293 cells with our constructs and with two additional plasmids coding for TLR4 and the accessory MD-2 protein. Very soon, we found out that our plasmid preparations were contaminated with LPS from E. coli (strain DH5α used for plasmid preparation), since there was no difference in the response between stimulated (with LPS) and unstimulated cells. To overcome this problem we decided to focus on TLR5 receptor which reacts upon contact with bacterial flagellin. Signal transfer through TLR5 does not depend on the presence of LPS.
Detection systems
In order to monitor the activation of the signaling pathway and function of our Parts we considered using several detection systems. We expected the synthesis of dominant negative protein (MyD88 or TRAF6), blocking of signaling pathway and consequently transcription termination of dominant negative protein. The constructs (parts) we prepared were designed for each detection system respectively.
Requests for the optimal detection system were:
- velocity;
- sensitivity;
- paralelization;
- optical signal would be preferred for the ease of data collection;
- in vivo and real time detection as a bonus;
- low price per assay.
On the basis of those requests we have decided for the following detection systems: flow cytometry, luminometry and ELISA. Members of the team in charge for each detection system are :
Constucts used in individual detection system and short description of detection systems:
Flow cytometry
Used composite parts: NFκB+dnMyD88-link-rLuc ([http://partsregistry.org/Part:BBa_J52014 BBa_J52014]).
Flow cytometry is a sophisticated method, which gives very precise and highly valuable results. The main advantage of this method is that measurements are done on individual cells, so the results are not the average value of the population. In our project we could detect cellular activation based on the phosphorylation cascade activation and anti-phosphoprotein antibodies. In our experiments we used primary antibodies (Ab) from mouse, which specifically detect only phosphorylated form of ERK kinase (pERK). The secondary antibodies used were anti-mouse Abs labelled with a fluorescence marker phycoerithrin (PE), which we can detect on flow cytometer. Cells were transfected with TLR5 or TLR4/MD-2 and with or without dnMyD88, which is under NF-κB promoter. On the second day the cells were stimulated with flagellin or LPS (depends on the selected TLR receptor). According to the literature phosphorylation of ERK can already be detected after 15 min. The amount of pERK kinase increases for additional 20 min and then it starts to decline. After 40 min no pERK is detected.
We used pulsed stimulation of cells. First we stimulated cells with 5 ng/ml of flagellin to activate the expression of dnMyD88. We repeated the stimulation with the second, stronger stimulus (50 ng/ml of flagellin) at different time intervals (2, 4, 6 and 8 hours after the first stimulation. 30 min after the second stimulus cells were fixed in 2% paraformaldehid, permeabilized with ice cold methanol and stained for pERK and secondary Ab labelled with PE.
Expected results: The activation of the signaling pathway also activates the expression of dominant negative protein MyD88, which should block the signaling, so we expect to detect decreased amount of pERK kinase after second stimulation in comparison to cells without of inducible dnMyD88.
Detection of transcriptional activation by luminometry
Used composite parts: NFκB+dnMyD88 ([http://partsregistry.org/Part:BBa_J52036 BBa_J52036]), NFκB+dnMyD88-link-rLuc ([http://partsregistry.org/Part:BBa_J52014 BBa_J52014]), NFκB+dnMyD88-link-rLuc-link-PEST191([http://partsregistry.org/Part:BBa_J52024 BBa_J52024]), NFκB+rLuc-link-PEST191([http://partsregistry.org/Part:BBa_J52023 BBa_J52023]), NFκB ([http://partsregistry.org/Part:BBa_J52010 BBa_J52010]), CMV-rLuc([http://partsregistry.org/Part:BBa_J52038 BBa_J52038]) and CMV+rLuc-link-PEST191([http://partsregistry.org/Part:BBa_J52039 BBa_J52039]).
Luminometry is a very used method to measure activation of signaling pathways. In many cases it replaced ELISA because it is much more sensitive. The basis of the method is the measurment of light, which is emited after substrate cleavage. We used two different enzymes; the firefly luciferase and the Renilla (sea pansy) luciferase can discriminate between their respective bioluminescent substrates and do not cross-activate, that is why we can measure the activity of both in one sample.
Dual luciferase assay
Dual luciferase assay includes two different luciferase reporter enzymes that are expressed simultaneously in each cell. Typically, the experimental reporter (in our case NFkB-fLuc) is correlated with the effect of specific experimental conditions, while the activity of the co-transfected "control" reporter gene (in our case CMV-rLuc) provides an internal control, which serves as the baseline response. Normalizing the experimental reporter gene to the activity of an internal control minimizes the variability caused by differences in cell viability and transfection efficiency. Thus, dual reporter assays allow more reliable interpretation of the experimental data by reducing extraneous influences.
Different experiments were conducted:
1. To be able to measure the activation of dnMyD88 expression after stimulation, we prepared dnMyD88 construct under NF-κB inducible promoter in the fusion with rLuc (construct NFkB+dnMyD88-link-rLuc). We transfected cells with TLR5 receptor plasmid and our biobrick construct. Cells were stimulated with flagellin. The expression of NFkB+dnMyD88-link-rLuc was measured in time intervals. We expect that amount of rLuc will increase in time only if cells are stimulated with a ligand.
2. CMV-rLuc-link-PEST191 biobrick was constructed to measure the kinetics of degradation of proteins in fusion with PEST. Cells were transfected with the construct. We found out that it takes four hours after the transfection to detect luciferase activity. To calculate half-life of rLuc and rLuc-PEST we transfected cells with either of those two constructs and incubated them to allow proteins to sythesize. New protein synthesis was inhibited by adding cycloheximide (inhibitor of protein synthesis in higher eucaryotes) and the activity of rLuc was measured in time intervals . Because the addition of cycloheximide stops the synthesis of new proteins there should be a decrease in rLuc activity due to protein degradation. A construct with PEST should decrease more rapidly as rLuc alone.
3. Cells were transfected with TLR5, NFkB-fLuc, CMV-rLuc and +/- NFkB+dnMyD88. To have the amount of NF-kB binding sites equal in all cells we added the same amount of plasmid containing only NF-κB inducible promotor to cells without NFkB+dnMyD88. The next day after the transfection cells were first stimulated with 5 ng/ml of flagellin to activate the signaling pathway and thus dnMyD88 expression. After 6 hours (time, which was measured as an optimal time for detection of the expression of dnMyD88) cells were restimulated with 50 ng/ml of flagellin. Activation of fLuc was measured in time intervals (0, 2, 4, and 6 hours after the second stimulation). The data were normalized for transfection efficiency by measuring rLuc. We expect that there will be a lower activation after the second stimulation in cells, which express dnMyD88.
ELISA for detection of transcription factors
Used composite parts: NFκB+dnMyD88 ([http://partsregistry.org/Part:BBa_J52036 BBa_J52036]) and NFκB+dnTRAF6-link–GFP ([http://partsregistry.org/Part:BBa_J52022 BBa_J52022]).
With ELISA detection system we are detecting free NF-κB transcription factor, which it was already shown that it is a good measurment of pathway activation. Active form of this protein is present after the activation of signaling pathway when protein is released from a complex with IkB inhibitor. Free NF-kB migrates into the nucleus where it acts as transcription activator. We are using high bind 96-well plates coated with [http://en.wikipedia.org/wiki/Streptavidin streptavidin](we prepare it ourselves). The probe for free NF-κB is biotinilated double stranded NF-κB binding sequence. The free NF-κB protein from whole cell lysate is captured by a probe, which biotin group binds to streptavidin coated on a plate. We detect NF-κB with primary mouse antibodies and secondary peroxidase-conjugated rabbit anti-mouse antibodies. Peroxidase is an enzyme, which cleaves a substrate that emits light, which we measure on luminometer.
Confocal microscopy
Microscope allows localization of signaling molecules within each individual cell, such as translocation of transcription factors, trafficking of proteins fused with reporter fluorescent proteins etc. Therefore this technique should be very usefull particularly for monitoring the expression of dnMyD88 inhibitory domain fused with fluorescent protein and the timecourse of its degradation. Therefore we designed Parts and several Devices, which included fusion with fluorescent protein:
[http://partsregistry.org/Part:BBa_J52021 BBa_J52021]
[http://partsregistry.org/Part:BBa_J52021 BBa_J52022]
[http://partsregistry.org/Part:BBa_J52021 BBa_J52026]
[http://partsregistry.org/Part:BBa_J52021 BBa_J52027]
[http://partsregistry.org/Part:BBa_J52021 BBa_J52040]
[http://partsregistry.org/Part:BBa_J52021 BBa_J52048].
Problems and Troubleshooting
Construction of Parts using bad oligos
We can only suggest to the other teams in the future to order oligos from a reliable and well established company (not necessarily the onw with the lowest price). When we repeatedly failed to produce PCR products we thought it could have been the beginner's bad luck, but when we determined a huge number of point mutations in the primer region even the mentors decided that we better avoid this company in the future (although they did offer us the replacement of oligos for free). There'll be no next time for this company in our lab.
Theory versus practise
We spent a lot of time trying to find an answer to the following question: "Why didn't our transformants grow on the LB-Tc plates?". First we accused our our tetracycline, than to the proper concentration, but at the end we found out that one of the previous year's parts was incorrectly registered. Now we know the importance of checking everything twice...
Real-time luminescence
We have attempted to monitor "real time" cell activation using luminescence and invested considerable amount of time and effort into this. We could not detectluminescence in vivo since coelentrazine (substrate of Renilla luciferaze) decays too quickly, while the luciferin could not penetrate the cells. We have also used the caged luciferin as well as its methy esther but in neither case did we suceed in detecting the luciferase in the living cells (several authors of published protocols were unavailable for comments :-( ).
Optimization of activation intervals
Much od lab work (more than initially planned) was devoted to the design of experiments to monitor cell activation. We had to find out the optimal concentration of inducers, the timespan between the first and second activation which gave the best difference, lenght of the experiment in which difference between genetically engineered and wild-type cells would be sufficiently prominent etc. Methods from the literature could not simply be adopted into our experimental procedure.
| Home | Background and Signaling Pathway | Project & Model | Results & Conclusions | Terms & References | Team members |
|---|