Missouri Western State University 2006
From 2006.igem.org
See [http://www.flickr.com/search/?q=igem+missouri photos] from the Ambassador visit on June 5-6, 2006.
Contents |
Students
• Marian L. Broderick [1]
• Adam D. Brown [2]
• Trevor L. Butner [3]
• Lane H. Heard [4]
• Eric L. Jessen [5]
• Kelly J. Malloy [6]
• Brad J. Ogden [7]
Faculty
• Todd Eckdahl [http://staff.missouriwestern.edu/~eckdahl/], Department of Biology, [8]
• Jeff Poet [http://staff.missouriwestern.edu/~poet/], Department of Computer Science, Mathematics, and Physics, [9]
Shipping Address: Todd Eckdahl, Biology Department, Missouri Western State University, 4525 Downs Drive, Saint Joseph, MO, 64507 [(816) 271-5873]
Assembly Plan
Math Stuff
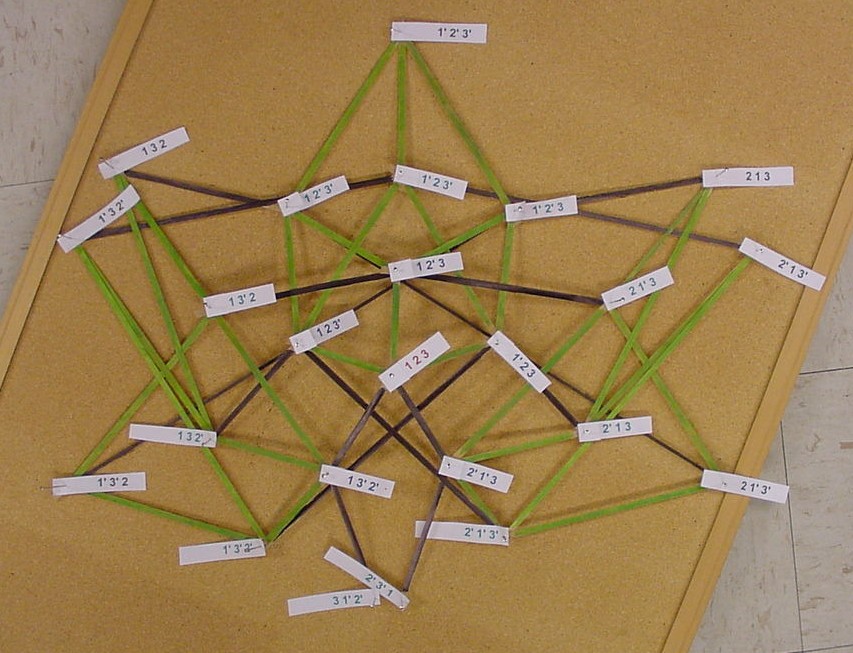
The picture below is of the half graph for the signed permutations for the 3-pancake problem. The full graph would have 48 vertices with each connected to six other vertices. The plan is to transfer this half graph onto the northern hemisphere of a globe with 123 as the north pole, five permutations on the Arctic Circle, eleven on the Tropic of Capricorn, and seven on the equator. An identical graph can be placed on the southern hemisphere such that each permutation occurs diametrically opposite its biological complement. In the picture below, green edges correspond to flips involving one pancake only and black edges correspond to a flip in which a double pancake is involved.
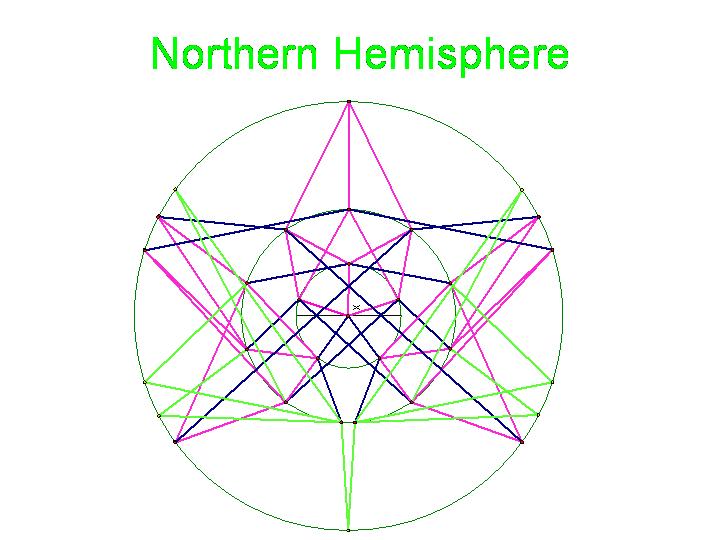
The graphic below was created using Geometer's SketchPad by Marian and shows the 24 vertices on the Northern Hemisphere, similar to the pushpin and rubber band manipulative above. The edges in this graph represent the relationships among the permutations. Pink edges correspond to the flip of a single pancake, black edges to the flip of a double, and green edges to vertices in the Southern Hemisphere. A 3-D model on a globe (okay, it is really a beachball) is in progress. Of curious note, doesn't this graph look similar to Da Vinci's Vintrium Man?
Publicity
MWSU Tower Topics [http://staff.missouriwestern.edu/~eckdahl/iGEM/Tower_Topics_7_2006.htm]
St. Joseph News-Press [http://staff.missouriwestern.edu/~eckdahl/iGEM/St_Joseph_NewsPress_7_2006.htm]
Research Links
OWW Protocols [http://openwetware.org/wiki/Protocols]
Source Plates [http://2006.igem.org/wiki/index.php?title=Biobrick_delivery&printable=yes]
Goodman et al. 1973-1979. The Pancake Problems [http://www.math.uiuc.edu/~west/openp/pancake.html]
Nanassy and Hughes. 1998. In Vivo Identification of Intermediate Stages of the DNA Inversion Reaction Catlyzed by the Salmonella Hin Recombinase [http://www.genetics.org/cgi/content/abstract/149/4/1649]
Lim et al. 1997. Hin-mediated Inversion on Positively Supercoiled DNA [http://www.jbc.org/cgi/content/full/272/29/18434]
Hughes et al. 1992. Sequence-specific interaction of the Salmonella Hin recombinase in both major and minor grooves of DNA [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=1628628]
Nanassy and Hughes. 2001. Hin Recombinase Mutants Functionally Disrupted in Interactions with Fis [http://jb.asm.org/cgi/content/full/183/1/28?view=long&pmid=11114897]
Lee et al. 1998. In Vivo Assay of Protein-Protein Interactions in Hin-Mediated DNA Inversion [http://jb.asm.org/cgi/content/abstract/180/22/5954]
Merickel et al. 1998. Communication between Hin recombinase and Fis regulatory subunits during coordinate activation of Hin-catalyzed site-specific DNA inversion [http://www.genesdev.org/cgi/content/abstract/12/17/2803]
Haykinson and Johnson. 1993. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=8508775]
Vasquez et al. 2004. Tangle Analysis of Gin Site-specific Recombination [http://math.berkeley.edu/~mariel/Vazquez_Gin04.pdf]
Merickel and Johnson. 2004. Topological analysis of Hin-catalysed DNA recombination in vivo and in vitro [http://www.blackwell-synergy.com/doi/abs/10.1046/j.1365-2958.2003.03890.x]
R.C. Johnson and M.F. Bruist. 1989. Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction [http://www.pubmedcentral.gov/picrender.fcgi?tool=pmcentrez&blobtype=pdf&artid=400990]
Lim and Simon. 1992. The Role of Negative Supercoiling in Hin-mediated Site-Specific Recombination[http://www.jbc.org/cgi/content/abstract/267/16/11176]
The Pancake Tossing World Record[http://www.recordholders.org/en/records/pancake.html]
Cool site for Breakfast [http://www.cut-the-knot.org/SimpleGames/Flipper.shtml]
iGEM iDeas
Powerpoint with two possible project ideas. [http://staff.missouriwestern.edu/~eckdahl/Research/Synthetic_Biology_2006/IGEMideas%2005-29-06.ppt]
Protocols
To Do List
Since I have severe separation anxiety, post what you did and how things turned out each day right here. Then I can offer suggestions and help plan the next steps.
Monday
Plasmid Prep
Vector Digestions
Develop Assembly Schedule
Tuesday
Trevor: We will have to make reversed RBS and reversed double terminator - design PCR primers for this the way we did for the reversed pAra on Friday and post the proposed sequences here.
cloning hix fragments
Wednesday
Making Reverse pARA
Thursday
Oligos should arrive for reverse RBS and TT (described on Assembly page). Do Reverse Parts with PCR
Pick four colonies off each of the transformation plates (except vector only plates) from yesterday and put into 5 ml LB+Amp at 37C overnight for plasmid preps tomorrow.
Friday
Do plasmid preps on overnight cultures
Measure plasmid yields with UV spec
Digest 200 ng (if that is not most of what we have) of plasmids with XbaI + SpeI and run on 12% PA
Start overnight cultures from transformations done on Thursday
Below are some questions from Jeff and some responses by Todd:
1. Can we double up on some constructions like Promoter(Forward) and Promoter(Reverse) by inserting them into a plasmid that already has a Hix site and getting a 50-50 split (which we can presumably separate out)?
I think it would be better to keep the two promoter orientations as separate constructions.
2. Could we start with a plasmid with a Hix site and insert several things at the same time and then sort them out later? (Example: could we put in a short promoter, a medium length RBS, and a long coding region at the same time and sort them out by length as to which connected? I think this is what you referred to as shotgun ligation when we were discussing the Traveling Salesman Problem.)
Again, I think that keeping things separate will be easier in the long run.
3. Should Hix be the LAST thing in any subconstruction so that we can create a version with HixL, one with HixR, and one with Hix C?
I guess this boils down to the question of how big of a library of parts do we want to create. Is it the case that every time we want to have a Hix site, we need one of each type of Hix? If we take this approach, then we can investigate any differences between the types of Hix sites, perhaps finding the most effective/efficient combination.
The way I see it, there will be four pancakes, each in two orientations, and each one will have a hixC on the right (3') end. That way, each could be used in modular fashion to make any of the four pancake constructs we want.
I think that for now we should head for the construction of parts that can be used in the two different one-pancake devices that will work in the four-pancake ones. I think we should concentrate for now on using hixC only. The problem can be subdivided into the following three devices: Hin recombinase producer, pancake flipping operon, and recombination enhancer.
Davidson Team: We believe we have Hix L, R and C cloned. We need to sequence these to verify. Also, we believe we have RE cloned. Again, we need to sequence this. We are still waiting for PCR primers for Hin. We will tackle this as soon as a pair shows up.
Davidson Team: We like Todd's idea of a part + HixC on the 3' end. The most efficient way to produce this is to use a plasmid with Hix already in it. Do NOT try to cut out a Hix and then clone it. It is too small (26 bp). So always clone the bigger piece into the HixC vector. However, for one pancake devices, we'll need to have Hix on bothsides of Promoter and none on the RBS+coding followed by a separate Terminator.
Davidson Team: We are working on the visuals and will add them to our wiki soon.
From Todd: Great work, Davidson Team! We need to continue to plan our assembly and divide and conquer. Jeff and our students are working on a schedule for assembly and there are also some thoughts at Develop Assembly Schedule. We are working on making reversed pAra this week and will also make plans to make reverse RBS and TT, which we will need eventually.