PLac Tet Pancake Plan
From 2006.igem.org
pBad Is a Weak Promoter
In our previous design, we aniticpated that a reverse RBS-RFP reporter would distinguish the biologically equivalent (1, 2) and (-2, -1) configurations of a pBad, TetR two-pancake stack. To test this, I placed the reverse RBS-RFP reporter to the left of (-2, -1). I also placed it to the left of (-1, -2) to see if the distance between the pBad promoter and the RFP would affect expression.
Only RFPrev-RBSrev-(-1, -2) confers weak expression of RFP (faint pink cell pellet), even after 0.2 - 2.0% arabinose induction in the absence of an extragenic copy of AraC (repressor of pBad). This result suggests that JM109 cells have endogenous AraC that represses pBad and that pBad is a weak promoter that requires close proximity to its coding region. Since one of our goals is to build long multi-coding sequence pancake stacks, pBad is a poor choice for this device (but may be useful in controlling Hin expression).
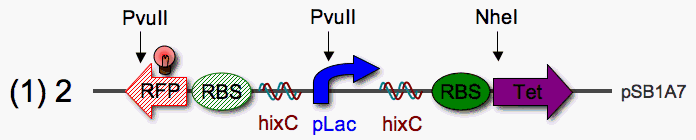
pLac Tet Pancake Assembly Plan
We've observed that pLac promotes strong expression of RBS-RFP, even in the absence of an inducer (IPTG). pLac may be better suited for our pancake stack device.
Restriction Mapping: PvuII sites occur once in the stationary RFP reporter and once in pLac. A PvuII digest can be used to check the orientation of pLac anywhere in the stack. Since PvuII and NheI share optimal buffers (Promega), a simple double digest could be used to check the orientation of the Tet pancake.
Construction, Step 1: