Princeton:Project Summary
From 2006.igem.org
PROGRAMMED DIFFERENTIATION OF MOUSE EMBRYONIC STEM CELLS USING ARTIFICIAL SIGNALING PATHWAYS
Our vision is to develop reliable techniques for programmed tissue generation in mammalian systems. Our iGEM2006 work encompasses artificial cell-cell signaling and signal processing, directed differentiation, pattern formation, modeling and precise cellular placement.
MAMMALIAN SYSTEMS
Senders and Receivers
In order to program mammalian cell-cell communication, we are designing a synthetic sender-receiver system based on the LuxI-LuxR pathway. This detection pathway is responsive to acyl homoserine lactone (AHL). Cells engineered to be senders synthesize and secrete AHL which rapidly diffuses across the cellular membrane. Receivers detect the presence of AHL and react, depending on the programmed response of the cells. In our multiplexed system, senders and receivers are separate populations of engineered cells. A multiplexer allows us to choose between two different cell fates using only one inducer. We can imagine needing to differentiate two different tissue types in close proximity to one another in vivo, for example muscle and nerve tissue. The toggle provides us with bistability after a transient application of inducer. This is advantageous in a living system because only a pulse of inducer is necessary for cells to differentiate into a programmed fate.
In our quorum sensing system, the engineered cells are both senders and receivers. Quorum sensing gives us an additional level of control over how many cells we have and when they will differentiate. Our two-step differentiation system adds an extra level of complexity and flexibility. From endoderm we have the potential to take the cells in many different directions depending on the genes expressed in the second programmed step. Here we choose to direct the cells toward pancreatic beta cells.
Multiplexed Cell Fate Determination
Embryonic Stem (ES) cells are pluripotent, i.e. they can differentiate into any tissue in the body. We have developed a 1:2 multiplexer which pushes the cells into one of two different cell fates depending on the presence or absence of AHL. We are using two known master cell fate regulators (CFRs): MyoD and Ngn1. The expression of MyoD is activated in the presence of AHL while Ngn1 is activated in the absence of AHL. The expression of MyoD pushes ES cells toward a muscle fate (myocyte formation) and expression of Ngn1 pushes ES cells toward a neuronal fate. We are incorporating cell deposition techniques in order to precisely place senders and create controlled pattern formation of different tissues. For this we use Laser Direct Write (LDW) of cells which can accurately deposit the cells in specified locations. We are also investigating the use of microfluidics to deliver inducing agents in controlled locations.
Sender-Receiver Directed Toggle Switch
We are in the process of constructing a biological toggle switch that is activated by cell-cell communication. This device will toggle between the expression of one of two CFR genes: MyoD and Ngn1. Each fate will be stable once the switch is flipped. We will measure the expression of these genes by using fluorescent reporters. Besides cell-cell communication via AHL, two other inducers are used to flip the toggle switch one way or another: IPTG and aTc.
Quorum Sensing Directed Beta Cell Differentiation
Our third system uses quorum sensing in order to direct two step differentiation of ES cells into insulin-producing pancreatic beta cells. The ES cells reach a critical density upon which genes responsive to AHL drive expression of endodermal cell fate regulators (Gata4 and Sox17). Once cells become endoderm, they produce a specific protein that drives the expression of other cell fate regulators (Pdx1 and Ngn3). Over expression of these CFRs will drive the cell lineage to pancreatic beta cells. Production of insulin produces red fluorescence.
Other CFRs, other systems
Our team concurrently devised a multiplexer system with neuronal CFRs which will determine whether cells will become motoneurons or interneurons. Pax6 is one of the CFRs involved in this system. Pax6 has the ability to change HeLa cells (cervical lineage) into cells with neuronal character.
MAMMALIAN BIOBRICKS
Team Princeton chose to create mammalian biobricks for iGEM2006.
The novel mammalian biobricks are used as components of systems designed by the iGEM2006 team as previously described. We focused on using these system components to differentiate mouse embryonic stem cells (and other mammalian cells) into tissues of our choosing. We have maintained the general concept of the modularity of the bacterial biobrick, although our choice to work with mammalian cells has necessitated the use of a unique design and construction method using lentiviruses. Lentiviral infection is much more efficient than transfection and the gene (or genes) of interest is stably integrated into the cellular genome.
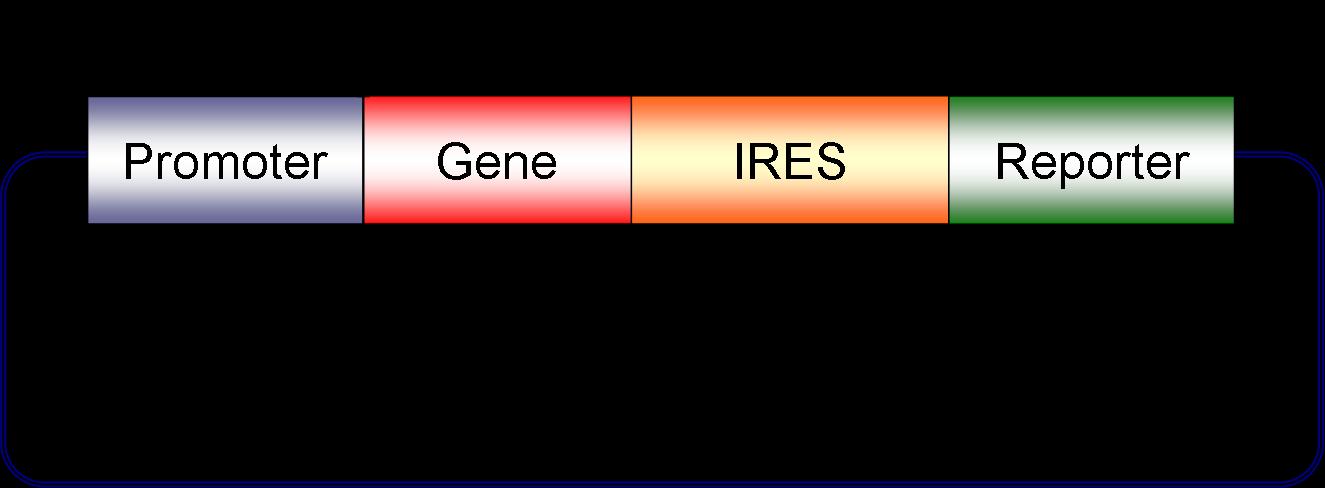
We have created a first version of a mammalian biobrick as shown below. Unique restriction sites surround the gene and the promoter allowing the user to quickly swap out these components. We are currently working on unique sites to modularize the reporter as well. Lentiviral biobricks are different from bacterial biobricks in that size limitations of the virus allow us to place only one promoter + gene + reporter combination in each viral particle. More than one promoter + gene + reporter cassette can be delievered to a given cell by coinfecting with multiple lentiviruses. Circuits are routinely comprised of multiple genes, promoters and reporters as discussed in our systems above. Coinfection of multiple engineered viruses allows us to easily place a full circuit inside one mammalian cell.
The Weiss and Lemischka labs are opening a central registry for mammalian biobricks. For inquiries on the mammalian registry, please contact Ron Weiss (rweiss@princeton.edu) or Ihor Lemischka (ilemisch@princeton.edu).