University of Texas 2006
From 2006.igem.org
m |
|||
| Line 1: | Line 1: | ||
The UT Austin team is: | The UT Austin team is: | ||
* Aaron Chevalier | * Aaron Chevalier | ||
| - | |||
* Eric Davidson | * Eric Davidson | ||
* [http://openwetware.org/wiki/User:Jeff_Tabor Jeff Tabor] | * [http://openwetware.org/wiki/User:Jeff_Tabor Jeff Tabor] | ||
| Line 16: | Line 15: | ||
For the 2004 Synthetic Biology Competition, our joint UT-Austin/UCSF team designed a genetically encoded edge detection device. In our system, a lawn of genetically identical precursor ''E.coli'' would be subjected to a light image, and only those cells at the light/dark boundary would become active and express a genetic reporter. | For the 2004 Synthetic Biology Competition, our joint UT-Austin/UCSF team designed a genetically encoded edge detection device. In our system, a lawn of genetically identical precursor ''E.coli'' would be subjected to a light image, and only those cells at the light/dark boundary would become active and express a genetic reporter. | ||
| + | An obvious hurdle in the implementation of this system was genetically encoding light detection in an easily manipulate-able and tractable system like ''E.coli''. To accomplish this, we used an incredible part engineered in the Voigt lab. This part, Cph8, ([http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5302 I15010]) is an engineered fusion between a cyanobacterial light sensing phytochrome (Cph1) and an ''E.coli'' transmembrane histidine kinase, (EnvZ). 660nm light causes an isomerization in the Cph1 domain of the chimera which strongly inactivates the histidine kinase acitity of EnvZ. When EnvZ is inhibited, a phosphorelay cascade which activates transcription from the ''OmpC'' promoter [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=3910 R0082]) and inhibits transcription from the ''OmpF'' promoter ([http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=3915 R0084]). We showed that when this system is expressed in ''E.coli'', it is possible to transform each cell on an agar surface into a decision making pixel capable of deciding whether it is in the light or dark. The community of cells is therefore capable of genetically reproducing a light image (figure to come). | ||
| + | This was the first step in engineering the edge detector, a massively parallel computation system that should be able to easily compute the edge of a complex light image; a very hard serial computation problem. We have now built the entire edge detection circuit ([http://partsregistry.org/Part:BBa_I15022 I15022]). | ||
| - | |||
| - | |||
| - | |||
| - | |||
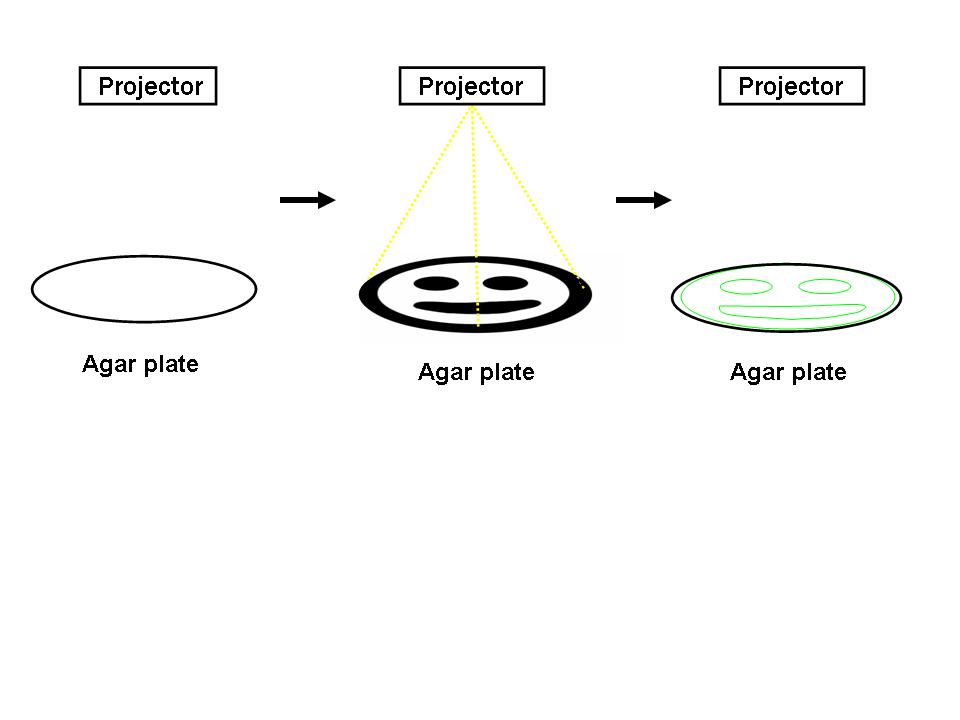
| - | + | [[Image:edge_detection.jpg|thumb|left|400px|Edge Detection. A light image is projected onto an agar plate of genetically identical ''E.coli''. Those cells at the light dark boundary express a reporter (green, right).]] | |
Revision as of 00:13, 1 November 2006
The UT Austin team is:
- Aaron Chevalier
- Eric Davidson
- [http://openwetware.org/wiki/User:Jeff_Tabor Jeff Tabor]
- Laura Lavery
- Matt Levy
- [http://www.mine-control.com/ Zack Booth Simpson]
Advisors:
- [http://ellingtonlab.org/ Andy Ellington]
- [http://polaris.icmb.utexas.edu/home.html Edward Marcotte]
Edge Detector
For the 2004 Synthetic Biology Competition, our joint UT-Austin/UCSF team designed a genetically encoded edge detection device. In our system, a lawn of genetically identical precursor E.coli would be subjected to a light image, and only those cells at the light/dark boundary would become active and express a genetic reporter.
An obvious hurdle in the implementation of this system was genetically encoding light detection in an easily manipulate-able and tractable system like E.coli. To accomplish this, we used an incredible part engineered in the Voigt lab. This part, Cph8, ([http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5302 I15010]) is an engineered fusion between a cyanobacterial light sensing phytochrome (Cph1) and an E.coli transmembrane histidine kinase, (EnvZ). 660nm light causes an isomerization in the Cph1 domain of the chimera which strongly inactivates the histidine kinase acitity of EnvZ. When EnvZ is inhibited, a phosphorelay cascade which activates transcription from the OmpC promoter [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=3910 R0082]) and inhibits transcription from the OmpF promoter ([http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=3915 R0084]). We showed that when this system is expressed in E.coli, it is possible to transform each cell on an agar surface into a decision making pixel capable of deciding whether it is in the light or dark. The community of cells is therefore capable of genetically reproducing a light image (figure to come).
This was the first step in engineering the edge detector, a massively parallel computation system that should be able to easily compute the edge of a complex light image; a very hard serial computation problem. We have now built the entire edge detection circuit ([http://partsregistry.org/Part:BBa_I15022 I15022]).