University of Texas 2006
From 2006.igem.org
The UT Austin team is:
- Aaron Chevalier
- Eric Davidson
- [http://openwetware.org/wiki/User:Jeff_Tabor Jeff Tabor]
- [http://openwetware.org/wiki/User:LLavery Laura Lavery]
- Matt Levy
- [http://www.mine-control.com/ Zack Booth Simpson]
- Bryan Kaehr
Advisors:
- [http://ellingtonlab.org/ Andy Ellington]
- [http://polaris.icmb.utexas.edu/home.html Edward Marcotte]
- [http://research.cm.utexas.edu/jshear Jason Shear]
Contents |
Previous work: Bacterial Photography
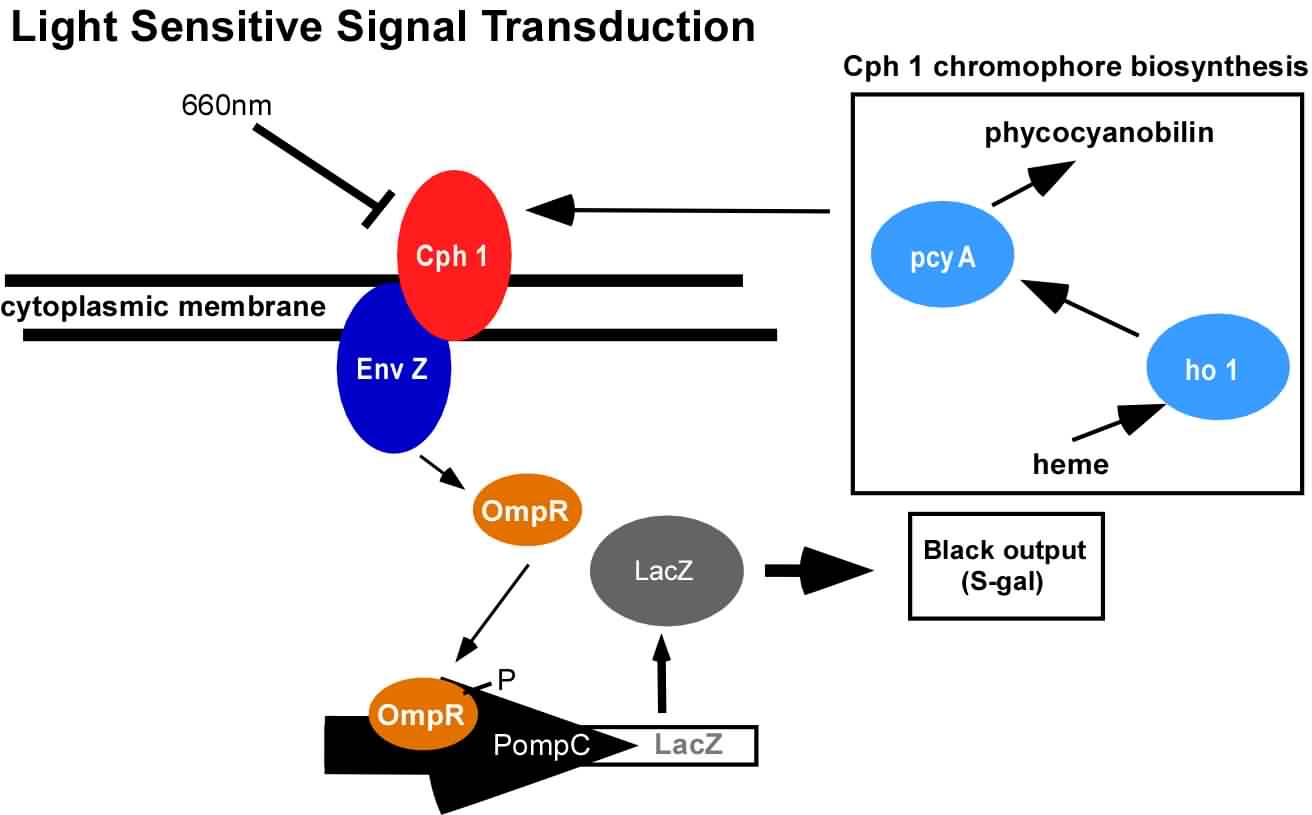
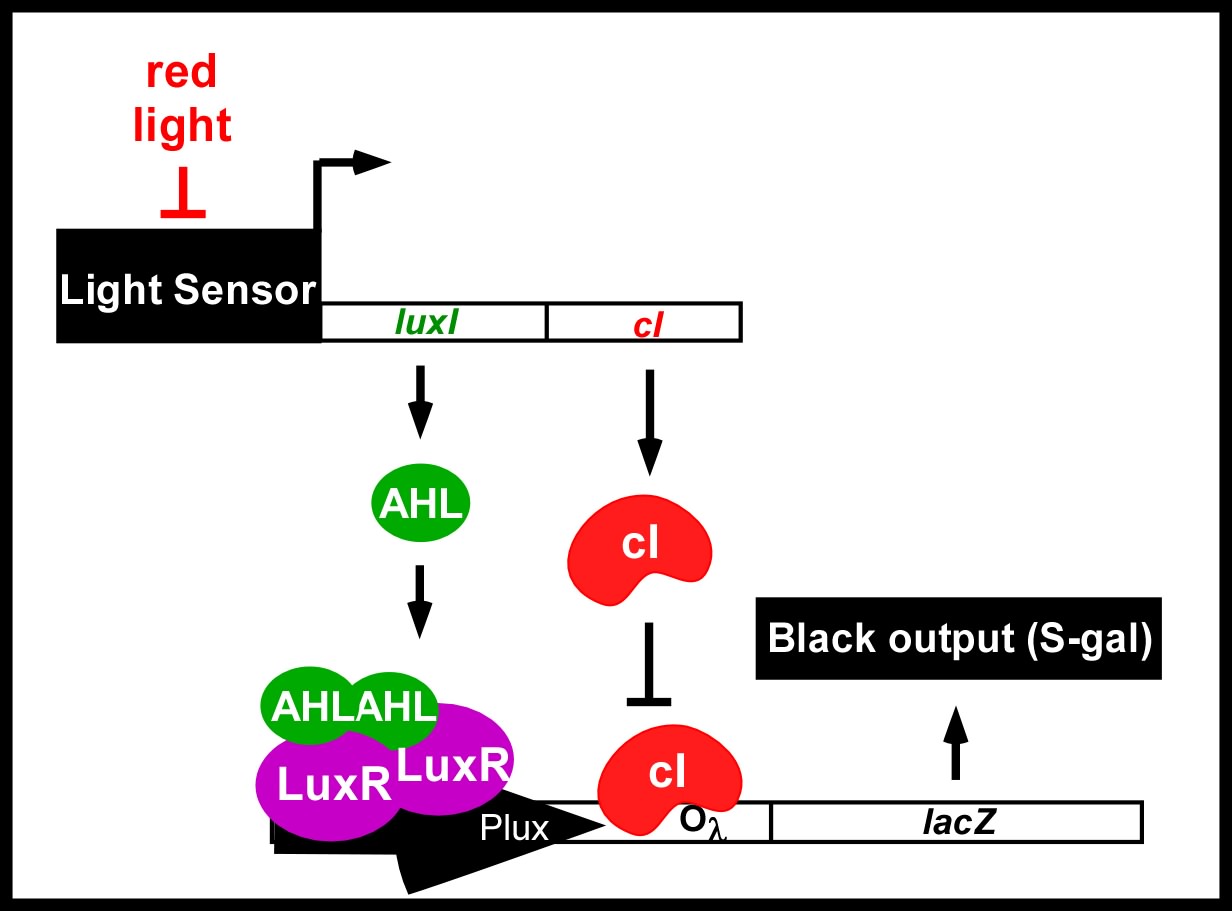
An obvious hurdle in the implementation of this system was genetically encoding light detection in the naturally blind E.coli. To accomplish this, we used a synthetic part engineered in the Voigt lab. This part, Cph8, ([http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5302 I15010]) is an engineered fusion between a cyanobacterial light sensing phytochrome (Cph1) and an E.coli transmembrane histidine kinase, (EnvZ). 660nm light causes an isomerization in the Cph1 domain of the chimera which inactivates the histidine kinase acitity of EnvZ. When EnvZ is inhibited, a phosphorelay cascade which activates transcription from the OmpC promoter [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=3910 R0082]) and inhibits transcription from the OmpF promoter ([http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=3915 R0084]). We then demonstrated that when this system is expressed in E.coli, it is possible to transform each cell on an agar surface into a decision making pixel capable of reporting whether it is in the light or dark. The community of cells is therefore capable of genetically reproducing a light image.
This year, we have added to our previous work with Cph8 by further characterizing its response under varying conditions, including the effect of osmolarity and different cell strains. Further, it has been known that the chromophore responds in opposite ways to light around 660 versus 735nm. Previously, light around 660nm was used to repress signal transduction. We now show that light around 735nm can be used to further activate signal transduction. If both lights are used simultaneously, an increase in contrast can be achieved. We have also characterized the one-plasmid biobrick vector containing the genes for both Cph8 and the biosynthesis of the chromophore.
Current work: Edge detector
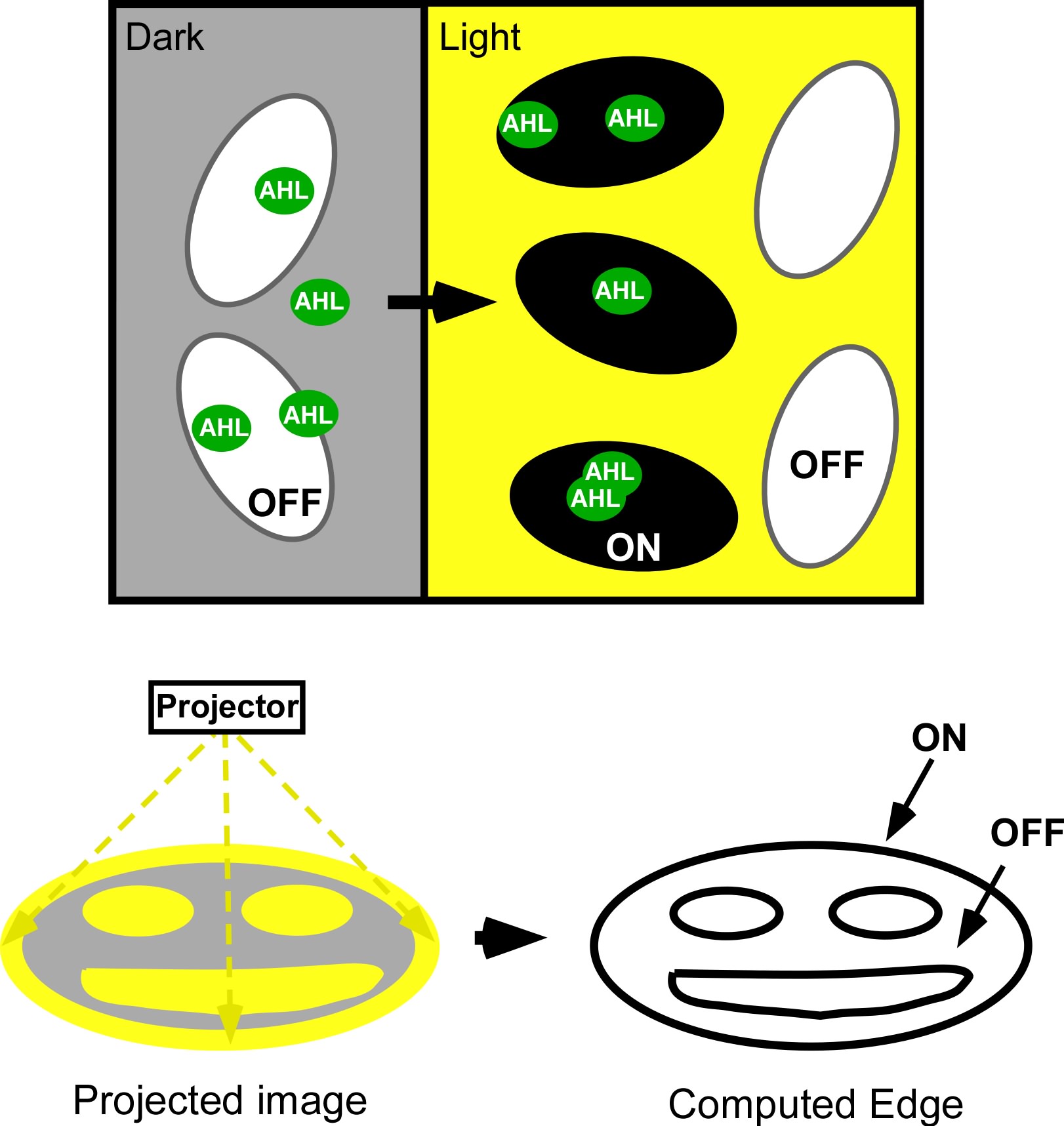
Light Controlled Chemotaxis
For future work, we would like to interface cellular motility with light detection. These bacteria could then be added to microstructures where bacteria power liquid flow through the structure in a light dependent manner. To accomplish this, we are currently cloning the CheR gene under the light-responsive OmpC promoter. Over expression of CheR results in methylation of the chemotaxis receptor which inhibits response to chemoattractants.
Photopolymerization
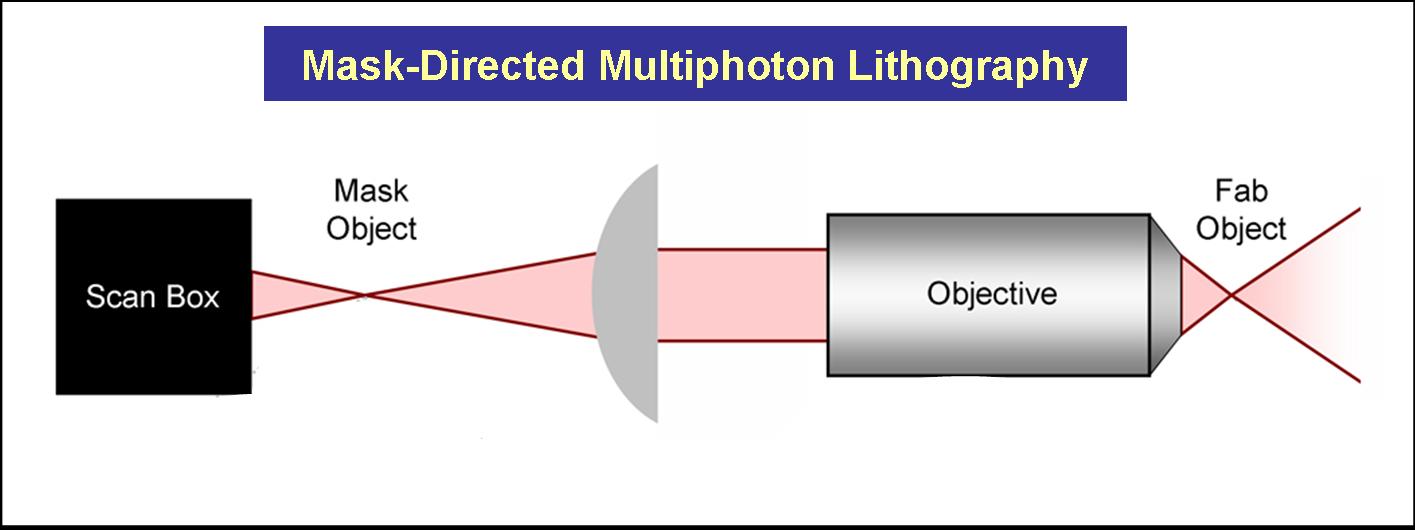
The Shear Lab at UT has developed methodologies that combine mask-directed lithography with multiphoton photo-crosslinking (MPP) to fabricate microstructures composed of cross-linked proteins. This approach allows for the rapid prototyping of complex micro-containers with feature sizes as small a 200 microns under fabrication conditions that are compatible with living cells.
Three dimensional structures can be built and used to trap and incubate small populations of cells, down to a single bacterium. Further, architectures can be proscribed to exploit cell motility and generate controlled, hydrodynamic microenvironments – a crucial step towards bio-powered pumps and mixers for microscale applications. These structures also have potential to be combined with engineered bacteria to control localized behavior of small bacterial populations or individuals.
Favorite Parts
- [http://partsregistry.org/Part:BBa_I15008 I15008]
- [http://partsregistry.org/Part:BBa_I15009 I15009]
- [http://partsregistry.org/Part:BBa_I15010 I15010]
- [http://partsregistry.org/Part:BBa_I15022 I15022]
Links
[http://www.utexas.edu/ UT Austin]
If you'd like to take your own pictures, check out the page on [http://openwetware.org/wiki/LightCannon how to build a light cannon]