NCBS Bangalore 2006
From 2006.igem.org
(→Lab notes) |
|||
| Line 168: | Line 168: | ||
State what the next step will be, based on these results. | State what the next step will be, based on these results. | ||
| + | |||
| + | |||
| + | '''Group II:''' | ||
| + | |||
| + | I am documenting the data that I presented in the Living Netwok Jamboree on June 3rd. | ||
| + | |||
| + | We are interested in synthetically synchronizing cell cycles. In the genetic construct we have used Pcya (adenylate cyclase promoter-that is known to oscillate in cell cycle dependant manner) as a core oscillator.LuxI tagged with cfp(+LVA) is expressed from this promoter.LuxI oscillations will result in AI(autoinducer) oscillations and this would result in oscillations of AI dependant promoter(yfp is expressed from this promoter). | ||
| + | CFP expression is readout for the core oscillator-Pcya and YFP is readout of the expression of the quorum sensing promoter0-Plux. | ||
Revision as of 08:58, 8 July 2006
Contents |
The Living Networks Wiki
December 12, 2024
The NCBS iGEM team grew out of an interdisciplinary workshop which ran through the spring of 2006. Participants included students, faculty, and industry researchers; physicists, biologists, computer scientists, and engineers; lawyers, educators, and entrepreneurs. Our discussions on genetic networks, synthetic biology, open-source ideas, and industrial applications, are documented on the workshop [http://www.ncbs.res.in/~faculty/ssb website].
Workshop participants
The team
Please feel free to change your titles. We'll order new business cards every week.
- Mukund: Chief Mentor and Brainstormer (thattai@ncbs.res.in)
- Adil: Chief Critic and Voice of Reason (adil@ncbs.res.in)
- Aparna: Chief Mind-Mapping Officer (aparna@ncbs.res.in)
- Ashesh: Head of Getting-Things-To-Work Department (ashesh@ncbs.res.in)
- Dhanya: Wild Ideas Consultant (dhanya@ncbs.res.in)
- Krithiga: Team Whip and Strategist (krithiga@ncbs.res.in)
- Ruchi: Head of Enthusiasm Department (mallik@ncbs.res.in)
- Sugat: Chief Technical Officer and Head of Big-Picture Department (sugat@ncbs.res.in)
The projects
Brainstorming notes
What should we work on?
- Group I:
- Develop a system by which Ecoli can sense two different colours. Red detection already exists, how can we pick up green independently? Can the two sensors be used to drive two different outputs? Try to couple these to X-Y chemotaxis. Problems: where can we quickly find a green sensor?
- Bacterial Fourier transform. Deliver some stimulus, e.g. light, in square pulses of a certain frequency. The output is the level of a reporter which can be correlated to frequency. Problems: how to make the amplitute of the output independent of the amplitute of the input signal, dependent only on the frequency?
- Bacterial signaling on wires and other patterns. Use a lawn of bacteria and some kind of template (e.g. a pattern of light) to activate certain cells. These cells can then talk to each other, transmitting a signal in defined patterns. Problems: for signalling around a ring, how to establish the initial asymmetry that makes transmission uni-directional?
- Group II:
- Using localized plasmids to detect the activity of a DNA-binding protein. (This is a research project in MT’s lab.) Introduce binding sites for CRP onto pole-localized plasmids; make a CRP-CFP fusion. When CRP is active, it will be pole localized; when it is inactive, it will be cytoplasmic. Problems: which pole-localized plasmid? How to ensure that additional binding sites and fusion proteins don’t perturb cell function?
- X-Y chemotaxis. We would like +/- control on two independent axes. This would require 4 different chemical gradients, two in each orthogonal direction. Suggestion: we could have default taxis in the ‘-' orientation, and active taxis in the ‘+’ orientation. Problems: how to make the cell ‘stand still’ in this configuration? Active feedback?
- Bacterial log book. If we have a single cell moving around a plate, can we reproduce its path? We would have to use DNA sequence rather than proteins, because the latter limits us to a few points. Use lambda insertion or transposon insertion; kill cells that return to the same point twice. Problems: The insertion mechanism is completely undefined.
- Synchronizing cell cycles. Use a cell cycle-dependent promoter (e.g. cya in Ecoli) to drive a cell-cell signal. Use the signal to modulate cell cycle in the receiver cell. This should allow synchronization of the population. Problems: How to modulate the cell cycle?
- Group III:
- Magnetic paintbox. Combine magnetotaxis with 3 colour detection to make a colour pattern generator. Problems: cannot reconstitute magnetotaxis in Ecoli.
- Synchronizing repressilators. This is based on a suggestion by Strogatz and Elowitz. Problems: the repressilator is an unreliable oscillator.
- An early warning chemotaxis system for UV damage. (This was suggested by Mrinalini Puranik.) A beam of UV light slowly moves over a bacterial lawn. Cells under UV suffer DNA damage and upregulate their repair mechanisms. Some of these cells actively move away from the light toward other parts of the plate, and trigger the repair response in neighboring cells. Problems: can we get unidirectional motion? How to sense UV damage? What pathways should we upregulate in receiver cells?
- Bacterial segregation by attractant/repellent strategy. Two populations of cells will be mixed. One synthesizes an attractant for its own kind; another synthesizes a repellent for the first kind. Together, this would cause cells to segregate, leading to Turing-like patterns. Suggestion: make use of molecules which can behave both as attractants and repellents depending on concentration (e.g. L-Lysine). Problems: we can make sure that different populations have distinct attractants, but we do not have control over repellents. Is a universal repellent sufficient to achieve the desired effect?
Each group has picked a single project upon which to focus. The task now is to think about intermediate goals, and chart out a research schedule for the coming month. The important aspect of each project is not the end result, but the design principles that will ensure a successful outcome. The question is: how to do control and validation experiments, and how to design intermediate constructs, that will maximize the chances of success. What we are developing here is not a product, but a process. The three projects span the range of construct complexity: one group requies a single regulated gene, while another is assembling a network of several genes and promoters. The simplest construct is most likely to do what we intend it to do; the more complex ones will require some coaxing. The projects also engage traditional biological questions to different extents: some aspects are interesting from a pure science perspective, while others are useful to engineers.
- Group I:
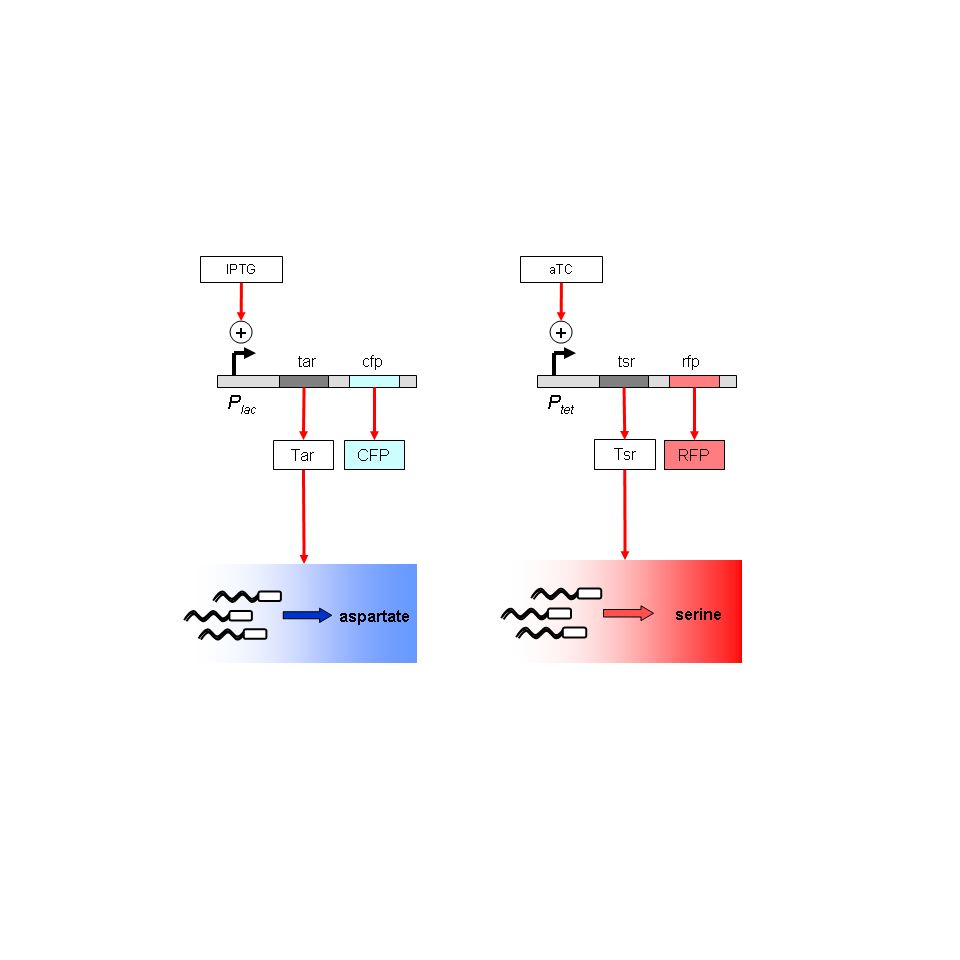
Project: Regulation of X-Y chemotaxis. Use two attractants whose receptors are inducible (serine with Tar, aspartate with Tsr). These define two orthogonal axes. Use a default attractant along a diagonal to establish '-' movement. Immediate goals: To establish substrate geometries upon which multiple reproducible, stable, quantitative chemical gradients can be established; to validate default chemotaxis in the absence of MCP receptors.
- Group II:
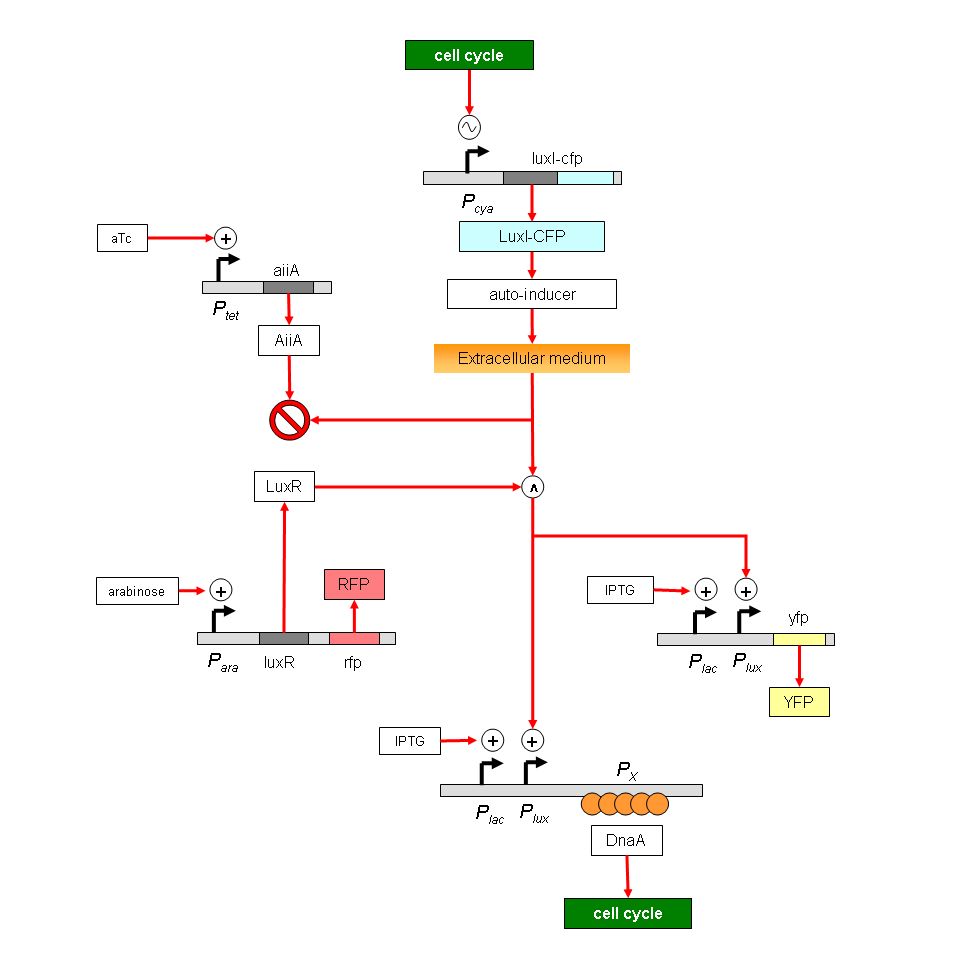
Project: Synchronization of bacterial cell cycles. Use a cell cycle-dependent promoter to drive a LuxI-LuxR based cell-cell signal. Use regulation of replication initiator DnaA to modulate cell cycle in receiver cells. Immediate goals: To determine if candidate promoters oscillate; to regulate DnaA levels.
- Group III:
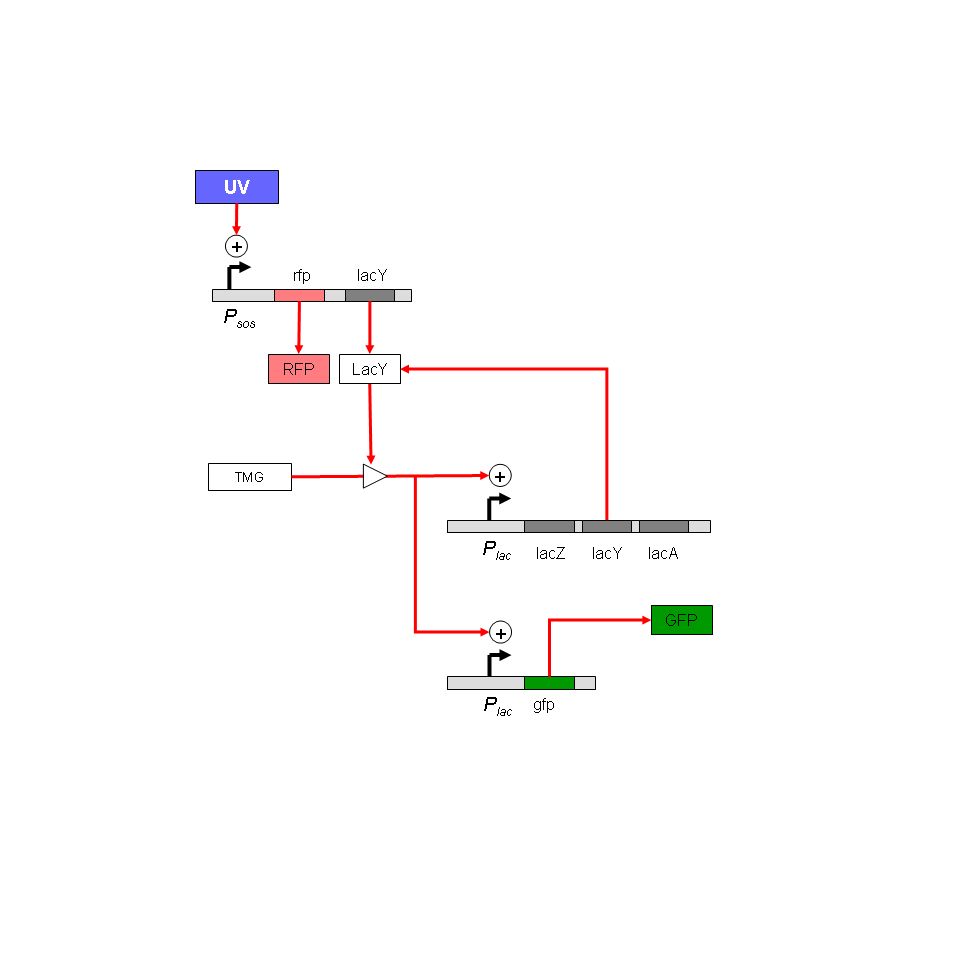
Project: Analysis of stress responses in biofilms. Connect stress response system to a lac based bistable GFP reporter. This allows transient expression at a stress-response promoter to trigger persistent expression of GFP. Immediate goals: To standardize biofilm assay; to design other experiments in which such a construct would be useful, especially using a cell-sorter.
Each group has submitted a DNA-level design for their construct. While these constructs are being synthesized, the groups should now work on measurement assays and apparatus.
- Group I:
Project: Regulation of X-Y chemotaxis. Ecoli strain UU1250 has 5 MCP-type chemotaxis receptors knocked out. The Tar and Tsr receptors (taxis towards Aspartate and Serine) will be put back on inducible promoters, Plac and Ptet. Chromosomal copies of lacI and tetR will be moved into strain UU1250 from strain Z1 by P1 transduction. Immediate goals: To test gradients in solid agar substrate; use various geometries and dyes to monitor time-dependence of gradients. Reservoirs and sinks seem a promising strategy.
- Group II:
Project: Synchronization of bacterial cell cycles. Complex construct is broken into simpler pieces to test the following aspects: (1) Does Pcya oscillate? Monitor LuxI-CFP fusion. (2) How much does oscillation attenuate: Use Plux and YFP. (3) How does DnaA sequestration affect replication? Use Plac to desequester DnaA. (4) How does DnaA affect Pcya? Use combination of constructs to test this. And so on. Immediate goals: To build a chemostat in which cell densities can be regulated, either by manual feedback or by limiting nutrients.
- Group III:
Project: Analysis of stress responses in biofilms. Stress-response monitor has been designed. Use RFP to monitor immediate SOS promoter activation, and GFP to monitor history of activation. Immediate goals: To standardize biofilm assay; to test TMG thresholds in solid medium.
Work has begun on developing assays and apparatus. Control experiments have been planned out.
- Group I:
Project: Regulation of X-Y chemotaxis. Chromosomal copies of lacI and tetR have been moved into strain UU1250 by phage transduction. The group has been exploring strategies for gradient formation using dyes in solid agar substrate. Immediate goals: Monitor time-dependence of agar chemical gradients. Use swarm-plate assays to characterize UU1250 and K12 chemotactic abilites. Fine-tune agar concentration and plate geometries so that gradients and cell motility are optimal.
- Group II:
Project: Synchronization of bacterial cell cycles. All measurements will be carried out in a chemostat, with succinate as the limiting nutrient. Strategies are being developed for initial cell synchronization. Immediate goals: Build and test the chemostat. Use FACS for cell synchronization.
- Group III:
Project: Analysis of stress responses in biofilms. Response of bistable Muk21 strain is being characterized in liquid and solid media. Strategies are being developed for systematic UV exposure. Immediate goals: To develop growth and measurement protocol for low cell densities.
- Group I:
Project: Regulation of X-Y chemotaxis. Behaviour of cells to light stimuli is being determined. The group has also developed several ways to generate sustained chemical gradients in soft agar. Swarm plate experiments have so far come up negative. Transformation of strain UU1250 with control plasmids is proving difficult. Immediate goals: To get the swarm plate assay working; to develop techniques allowing the use of 0.25% agar (blotting paper to prevent condensation; trying agarose instead of agar); to try transformation protocol in less stringent conditions; to generate gradients in liquid media.
- Group II:
Project: Synchronization of bacterial cell cycles. The group has built a chemostat using succinate as the limiting nutrient. The chemostat is able to maintain constant cell densities for several hours. They have attempted to synchronization of cell cycles using the cell-sorter; however, neither fluorescence nor scattering seem to correlate with cell size. Immediate goals: To try alternate strategies obtaining synchronized cells; to design a ‘mini-chemostat’ for maintaining cell densities and cell-cell communication on the microscope stage.
- Group III:
Project: Analysis of stress responses in biofilms. The group has been standardizing the response of Muk21 cells to TMG stimulus. They have measured growth curves and standardized protocols for low-cell-density experiments. Immediate goals: To test two different techniques for growing biofilms; to quantitate the UV-exposure procedure.
- Group I:
Project: Regulation of X-Y chemotaxis. Chemical gradients have been developed using a combination of low- and high-concentration agar. These gradients are stable over days. Methods of setting up gradients in liquid media are being tested. Immediate goals: To reduce the scale of the apparatus, so that the entire system can be imaged, and the timescales required to test chemtactic ability are reduced.
- Group II:
Project: Synchronization of bacterial cell cycles. A new strategy is being developed to synchronize cells using the cell-sorter. The process involves two successive sorting procedures: the first to eliminate noise in fluorescence intensities, and the second to detect recently divided cells. The Px-promoter construct is ready. Immediate goals: To test DnaA-binding ability of Px promoter; to use Px in a cell synchronization strategy.
- Group III:
Project: Analysis of bacterial stress responses and early warning systems. The group has attemped to use a flow-cytometer to distinguish induced and uninduced Muk21 cells. However, the fluorescence intensity is too low to enable detection. Immediate goals: To understand all inducers of the SOS system; to verify bistability of Muk21 strain using fluorescence microscopy.
This week, ask how quantitative modelling can help in the design process. Each group was asked to outline a few key questions which could be addressed using models of various kinds. There are some issues to keep in mind before taking the time to develop a model: 1. Is there enough data to constrain the space of possible models? Is enough quantitative information available for parameter values to be determined? 2. Will the results of the model aid in the design of constructs, or in the design of experiments to test the constructs? Can a model be used to eliminate certain scenarios, so that effort can be concentrated on key questions?
- Group I:
Project: Regulation of X-Y chemotaxis.
- Biophysical diffusion model of chemical gradients in various geometries. The model will be used to infer the profile of amino-acid gradients based on measurements made with fluorescent dyes, accounting for mass and mobility differences. It will provide information about stability and equilibration timescales.
- Bacterial chemotaxis at the single-cell level. The model will be based on those which exist in the literature, and include the effect of runs and tumbles of individual cells.
- Induction of chemotaxis receptors. The model will include protein synthesis and degradation, and will predict how long the system will take to switch from one attractant to another. It will include the effect of genetic noise and plasmid number fluctuations. Ultimately, all these models will be combined into a single predictive platform.
- Group II:
Project: Synchronization of bacterial cell cycles.
- Degradation of peak-to-trough ratio of oscillatory signal. The oscillatory signal from the cya promoter will lose quality as it is passed through successive signalling stages. However, there are certain protein concentrations (AII and LuxR, for example) whose levels can be regulated in order to minimize this problem. The model will be used to determine optimal protein concentrations.
- Biophysical analysis of de-sequestration by RNA polymerase. A detailed model will be constructed incorporating known binding energies and polymerase forces, to determine whether a moving polymerase can knock off a DNA binding protein from its binding site.
- Group III:
Project: Analysis of bacterial stress responses and early warning systems.
- Modelling the LacY induction strategy of Muk21. The system is essentially a pulse discriminator: short induction pulses will not cause GFP synthesis, longer pulses will cause sustained synthesis. The model will predict the tightness of this response, accounting for plasmid number fluctuations.
- Using Muk21 as a sensitive lacY sensor. Based on the results of step (1), can Muk21 cells be used to measure small diferences in LacY levels?
Lab notes
Please add all experimental data here.
Summarize the goal of the experiment, and the results.
State what the next step will be, based on these results.
Group II:
I am documenting the data that I presented in the Living Netwok Jamboree on June 3rd.
We are interested in synthetically synchronizing cell cycles. In the genetic construct we have used Pcya (adenylate cyclase promoter-that is known to oscillate in cell cycle dependant manner) as a core oscillator.LuxI tagged with cfp(+LVA) is expressed from this promoter.LuxI oscillations will result in AI(autoinducer) oscillations and this would result in oscillations of AI dependant promoter(yfp is expressed from this promoter). CFP expression is readout for the core oscillator-Pcya and YFP is readout of the expression of the quorum sensing promoter0-Plux.