Penn StateProjectDes
From 2006.igem.org
(→'''Preliminary Cell Designs''') |
(→'''Cell Designs''') |
||
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
== '''Bacterial Relay Race''' == | == '''Bacterial Relay Race''' == | ||
| - | ''Pennsylvania State University'' | + | ''The Pennsylvania State University'' |
''iGEM 2005'' | ''iGEM 2005'' | ||
| - | == | + | ==Conception== |
| - | + | The idea for our project grew out of one for a "bacterial maze," in which bacteria would use logic to make their way through a microfabricated labrynth. This seemed slightly too difficult, so we linearized the the concept and added transfer of a signal; the idea was then dubbed a "bacterial relay race." | |
| - | + | ||
| - | + | ||
| - | + | As in a conventional relay race, the signal is to "go," or induce motility of a latter stage participant. This is accomplished by passing a baton. In our case, the participants are E. coli, and the baton is a quorum sensing molecule, 3OC6HSL (we have another strategy that utilizes conjugation rather than quorum sensing to mediate the signal). | |
| - | + | ||
| - | + | In addition to passing the signal, though, the first participant must stop. We explored this option, but settled instead on terminating the first participant. In our design we really do kill the messanger. | |
| - | == ''' | + | The second leg of the race must remember to keep running; this is easy because they are gripping a baton. Our HSL baton, however, degrades over time. Thus, it is important to for the second cell to retain memory of the signal event. We incorperate this feature into our design by introducing a switch, which, barring sufficient stochastic fluxuations, should remain in the off state until prompted into the on state by the signal. |
| - | + | ||
| - | [[Image: | + | Under control of the switch is a GFP reporter and MotB, the latter a protein linking the torque machinery of the flagellum to the cell wall. By placing our system in a motility-comprimised strain (i.e. MotB K/O) and complementing it with the functional copy of the MotB listed above, we are able to induce motility. |
| + | |||
| + | |||
| + | How to Engineer it? | ||
| + | |||
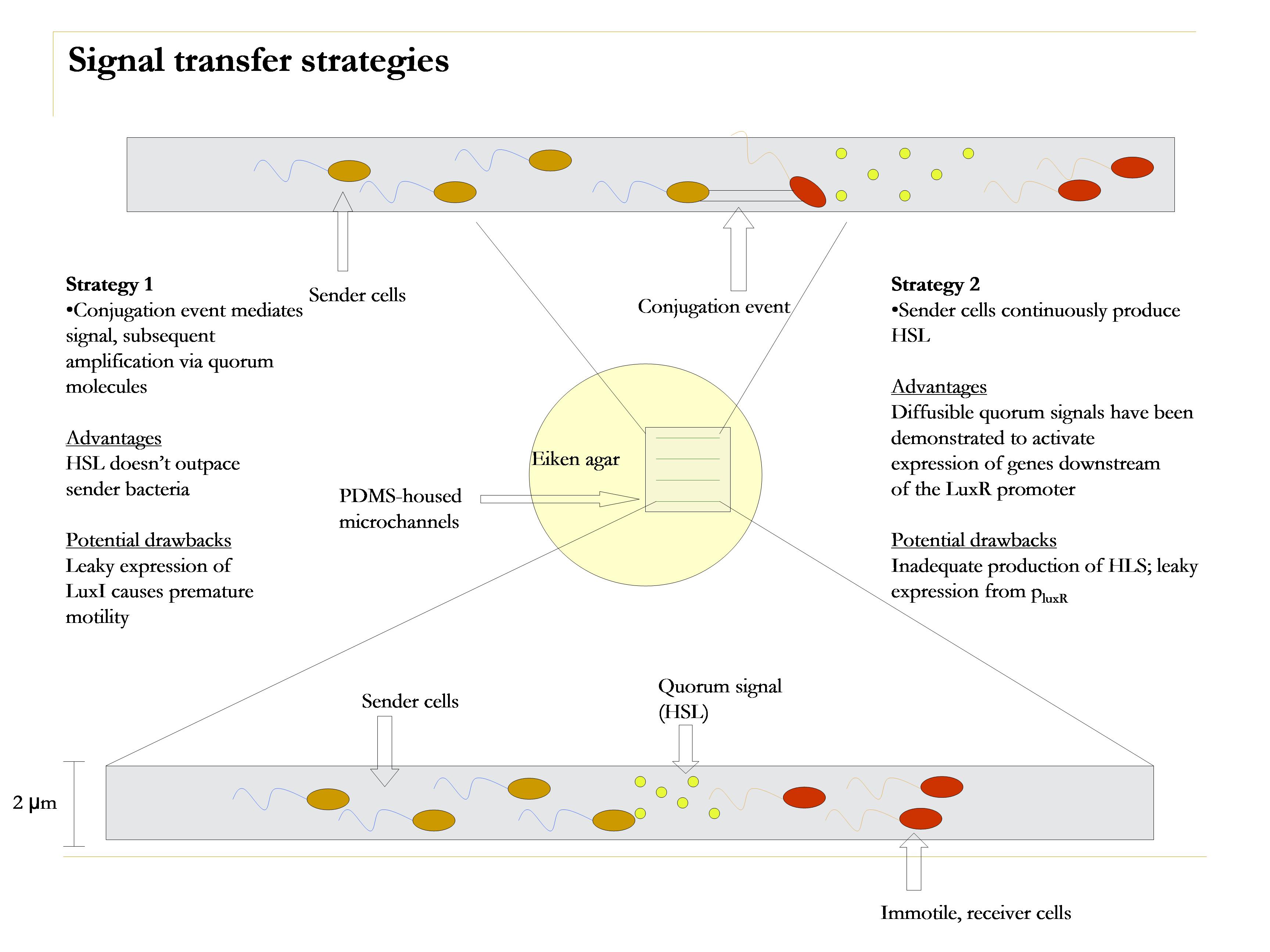
| + | [[Image:Strategies.jpg|thumb|left|250px|caption.]] | ||
| + | |||
| + | |||
| + | |||
| + | Parts assembled: | ||
| + | |||
| + | R0062+E0240 | ||
| + | |||
| + | R0062+P0112 | ||
| + | |||
| + | R0010+E0240 | ||
| + | |||
| + | Q04510+P0112 | ||
| + | |||
| + | Q04510+E0240 | ||
| + | |||
| + | R0010+S03271 | ||
| + | |||
| + | R0062+P0112+R0010+E0240 | ||
| + | |||
| + | R0010+Q04510+P0112 | ||
| + | |||
| + | == '''Cell Designs''' == | ||
| + | |||
| + | |||
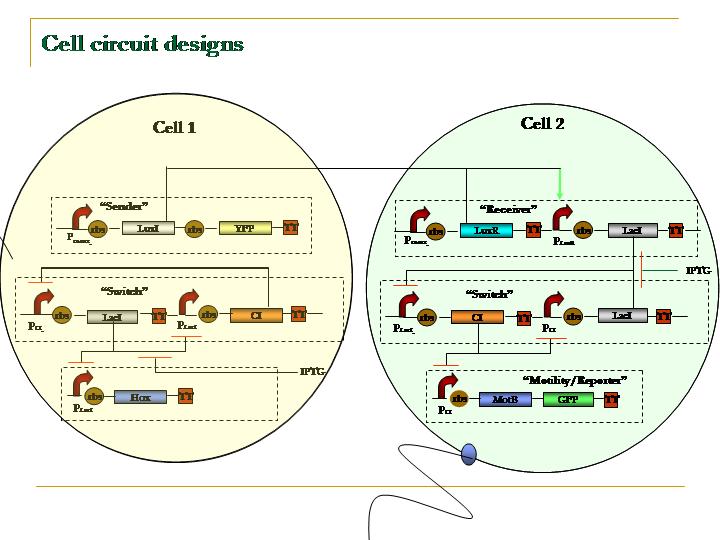
| + | [[Image:PSUcircuit.jpg]] | ||
| + | |||
| + | == presentation == | ||
Latest revision as of 17:10, 8 November 2005
Contents |
Bacterial Relay Race
The Pennsylvania State University iGEM 2005
Conception
The idea for our project grew out of one for a "bacterial maze," in which bacteria would use logic to make their way through a microfabricated labrynth. This seemed slightly too difficult, so we linearized the the concept and added transfer of a signal; the idea was then dubbed a "bacterial relay race."
As in a conventional relay race, the signal is to "go," or induce motility of a latter stage participant. This is accomplished by passing a baton. In our case, the participants are E. coli, and the baton is a quorum sensing molecule, 3OC6HSL (we have another strategy that utilizes conjugation rather than quorum sensing to mediate the signal).
In addition to passing the signal, though, the first participant must stop. We explored this option, but settled instead on terminating the first participant. In our design we really do kill the messanger.
The second leg of the race must remember to keep running; this is easy because they are gripping a baton. Our HSL baton, however, degrades over time. Thus, it is important to for the second cell to retain memory of the signal event. We incorperate this feature into our design by introducing a switch, which, barring sufficient stochastic fluxuations, should remain in the off state until prompted into the on state by the signal.
Under control of the switch is a GFP reporter and MotB, the latter a protein linking the torque machinery of the flagellum to the cell wall. By placing our system in a motility-comprimised strain (i.e. MotB K/O) and complementing it with the functional copy of the MotB listed above, we are able to induce motility.
How to Engineer it?
Parts assembled:
R0062+E0240
R0062+P0112
R0010+E0240
Q04510+P0112
Q04510+E0240
R0010+S03271
R0062+P0112+R0010+E0240
R0010+Q04510+P0112