Arsenic Biosensor
From 2006.igem.org
| (88 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
| - | == | + | == The worst mass poisoning the world has ever known == |
| - | This | + | [[Image:bangladesh.jpg|400px|thumb|right|This photo is the courtesy of [http://www.wateraid.org WaterAid] / Liba Taylor]] |
| + | |||
| + | It is estimated that at this very moment up to 100 million people across the world are being poisoned due to the presence of arsenic in their drinking water. One in a hundred of these people will develop a fatal cancer as a result, and countless others will get ill and suffer as the poison slowly damages their bodies. | ||
| + | |||
| + | This problem is greatest in one of the world’s poorest countries – Bangladesh. Because of the combination of regular yearly floods and poor sanitation, the available fresh water is swarming with harmful diseases. In an attempt to alleviate this problem, a large number of tube-wells were dug all across the country in order to provide people with clean water. However, in a cruel twist of fate, a lot of these wells were dug through a sedimentary layer containing arsenic. | ||
| + | |||
| + | Arsenic wasn't a standard thing to test for at the time, and by the time the first signs of the poisoning emerged, it was already too late – out of the estimated 10 million wells dug by now, at least 1 in 4, and possibly almost as many as a half of these wells contain water with harmful levels of arsenic. The organisations working in present day Bangladesh do carry out routine tests for arsenic in new wells, and arsenic mitigation efforts are being carried out, but the immense scale of the problem means these are basically just a drop in the ocean. | ||
| + | |||
| + | So the choice facing a poor rural family in Bangladesh is one between a quick death from disease if they drink the water from the rivers, or the potential of slow death from poisoning if they drink the water from the wells. The problem is that they don’t know which wells are poisoned, and because the effects of arsenic are slow and cumulative, by the time they find out, it’s already too late. | ||
| + | |||
| + | 1 in 4 wells. Would you play Russian roulette with your children’s lives? | ||
| + | |||
| + | |||
| + | == The project == | ||
| + | |||
| + | Scientists around the world have been working to change this. The distribution of arsenic in the wells is quite irregular, so if a cheap, easy-to-use arsenic detector was to become available, all the wells could be tested, and safe ones could be found – something that is currently financially unfeasible because of the instruments and expertise required. | ||
| + | |||
| + | A lot of research has been done into arsenic biosensors already, and some have already been created, mostly using fluorescence. The components are available, there are gene promoters that respond to the presence of arsenic, and various different outputs are possible. The next stage is really more of an engineering question: how do you come up with an optimal design that’s easy to use in the field? | ||
| + | |||
| + | Our suggested project uses a pH change as a signal for the presence of arsenic, improving upon previous designs because the change is easily detectable using a pH meter, or even just a couple of drops of indicator. It is also potentially possible to make the system detect a large range of arsenic concentrations, because the change in pH is gradual. | ||
| + | |||
| + | Not only is such a device both practical and marketable, it is ethically imperative to produce it. Using the cutting edge of modern science, it really is possible to change the world. | ||
== Possible mechanism 1 - the bare minimum == | == Possible mechanism 1 - the bare minimum == | ||
| + | |||
| + | === Outline === | ||
INPUT [Arsenate/Arsenite] -> Detector -> Reporter -> OUTPUT [H+] | INPUT [Arsenate/Arsenite] -> Detector -> Reporter -> OUTPUT [H+] | ||
The arsR gene codes for a repressor which binds to the arsenic promoter in the absence of arsenate or arsenite. These two genes can be linked to the lacZ gene, in order to place lactose metabolism under the control of an arsenic activated promoter. | The arsR gene codes for a repressor which binds to the arsenic promoter in the absence of arsenate or arsenite. These two genes can be linked to the lacZ gene, in order to place lactose metabolism under the control of an arsenic activated promoter. | ||
| - | |||
| - | |||
The ability to metabolise lactose should result in the acidification of the medium, and an output of H+ ions, giving a pH response to the input of arsenate/arsenite molecules. | The ability to metabolise lactose should result in the acidification of the medium, and an output of H+ ions, giving a pH response to the input of arsenate/arsenite molecules. | ||
| Line 25: | Line 46: | ||
It should be possible to obtain a curve for pH vs. arsenic concentration, where a range of pH values representing an unsafe level, a safe level, or an intermediate level of arsenic is present can be elucidated after calibrating the biosensor with known concentrations of arsenic. | It should be possible to obtain a curve for pH vs. arsenic concentration, where a range of pH values representing an unsafe level, a safe level, or an intermediate level of arsenic is present can be elucidated after calibrating the biosensor with known concentrations of arsenic. | ||
| + | '''Parts needed''': | ||
| + | Arsenic binding repressor protein | ||
| + | Arsenic repressor protein responsive promoter | ||
| + | lacZ protein coding gene | ||
| + | |||
| + | |||
| + | === Preliminary experiments === | ||
| + | |||
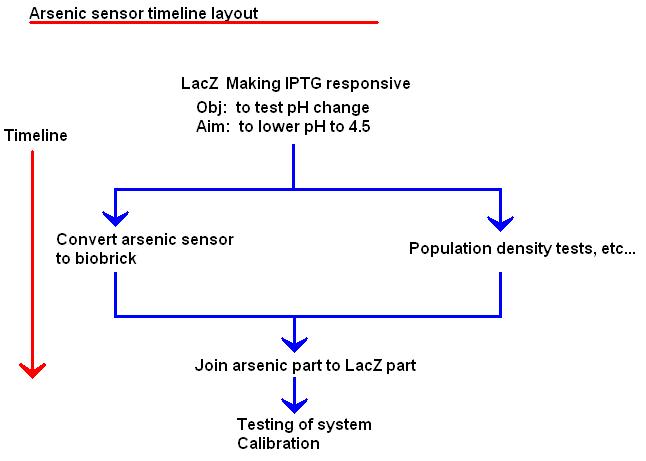
| + | [[Image:Arsenic_sensor_timeline_layout_1.JPG|Arsenic sensor timeline layout]] | ||
| + | |||
| + | *How fast (if at all) E. coli change the pH of the medium when the lacZ gene is activated. | ||
| + | |||
| + | *Playing with the lacZ gene attached to a promoter we can easily activate in the lab, so that we can see how the pH changes with regards to bacterial density, quantity of lactose present, growth stage the bacteria are in, etc. | ||
| + | |||
| + | *Testing the response of the reporter genes when ArsR is bound by arsenic | ||
| + | |||
| + | |||
| + | A lot of this project will be calibrating the sensor, finding the time to reach a steady state, finding out which amount of cells is ideal for reaching the steady state which gives distinct results for each pH range, and finding out which pH range represents which concentration of arsenic. It would be ideal to have the threshold for the legal limit of arsenic at the point of the curve which gives the most accurate result. | ||
== Possible mechanism 2 - more ambitious == | == Possible mechanism 2 - more ambitious == | ||
| + | |||
| + | ==='''<font color='blue'>Get the powerpoint animation on how the system works [http://2006.igem.org/Image:Presentation_only_1.ppt here!]</font>'''=== | ||
| + | |||
| + | |||
| + | [[Image: Model_Diagram2.JPG|400px|thumb|right|Arsenic Model Diagram]] | ||
After discussion, it was decided that it would be unfeasible to try and engineer a sensitive enough analogue output just using the pH - instead, a set of a couple of defined 'stepped' outputs would be preferable. | After discussion, it was decided that it would be unfeasible to try and engineer a sensitive enough analogue output just using the pH - instead, a set of a couple of defined 'stepped' outputs would be preferable. | ||
| Line 32: | Line 76: | ||
In order to increase the range that is indicated by the biosensor, the following mechanism might be worth considering. | In order to increase the range that is indicated by the biosensor, the following mechanism might be worth considering. | ||
| - | + | '''Sample:''' Water sample + Lactose + Urea + Bacterial culture | |
| + | The mechanism involves 4 'devices' inside the bacteria. | ||
| + | '''Device 1:''' The lac operon regulatory region is connected to the LuxR coding gene. | ||
| + | '''Device 2:''' The LuxR/cI responsive promoter is connected to the gene for urease which, if activated, catalyses the conversion of urea into ammonia and carbon dioxide, raising the pH. | ||
| + | |||
| + | '''Device 3:''' A promoter that's regulated by a repressor very sensitive to arsenic. Thus anything above 5 ppb of arsenic would bind to the repressor and enable gene expression. This promoter is connected to a gene that expresses the lambda cI molecule, thus keeping the pH at neutral. | ||
| + | |||
| + | '''Device 4:''' A promoter regulated by a repressor that's somewhat less sensitive to arsenic, so only switches on at, say, 20 ppb. Connected to lacZ, as detailed above in Possible mechanism 1. The LacZ breaks down the lactose present, shutting down device 1. | ||
| + | |||
| + | |||
| + | So, if all goes according to plan, we would get the following results: | ||
| + | |||
| + | '''If no arsenic is present:''' The lac operon in Device 1 detects the presence of lactose that's been put in, activates the expression of the genes behind it, and urease catalyses the reaction that raises the pH of the surrounding solution. So the pH with no arsenic present is around 9-10. | ||
| + | |||
| + | '''If 5 ppb arsenic is present:''' The arsenic activates Device 2, which produces a repressor that deactivates Device 1. No enzyme is produced, and the pH remains neutral (or maybe slightly higher than neutral because urea is present, but there's still a distinguishable difference). 5ppb arsenic = pH 7 | ||
| + | |||
| + | '''If 20 ppb arsenic is present:''' The arsenic activates Device 3, which produces lacZ, which digests the lactose in the solution and turns it acidic. Device 2 is still switched on and Device 1 is still switched off accordingly, so there are no problems with the acid and the alkali neutralising one another. 20ppb arsenic = pH 4.5 | ||
| + | |||
| + | The figures for the concentrations and the pH are basically currently just made up, it would take weeks of calibration in the lab to tune everything properly and see what's feasible. Whether the E. coli survives the given range of pH or not is basically irrelevant, as each infusion of bacteria would be single use, and neither the alkali nor the acid will be produced until the bacteria are already sitting in the test sample. | ||
| + | |||
| + | === Parts required === | ||
| + | |||
| + | |||
| + | '''Device 1:''' | ||
| + | |||
| + | *RBS - BBa_B0030 | ||
| + | *Promoter constitutively switched on. Part BBa_I14032, promoter P(Lac) IQ | ||
| + | *LacI gene: present in the registry, BBa_C0012. Has an LVA degradation tail attached, not sure if we need that. Doesn’t say it’s been found to be working, maybe we could make our own? | ||
| + | *Terminator: say BBa_B0015? It seems to be the most efficient one that works. We want it to be efficient, right? | ||
| + | *LacI regulated promoter: BBa_R0011, extensively tested and found to work. Mutated from a Lambda phage. Should be fine to use this one. | ||
| + | *LuxR coding gene - part BBa_C0062, seems like a good part, but there's no reports on whether it works. | ||
| + | *Terminator: BBa_B0015 | ||
| + | |||
| + | |||
| + | '''Device 2:''' | ||
| + | |||
| + | *RBS - BBa_B0030 | ||
| + | *Part BBa_R0065 - lambda cI and luxR regulated promoter | ||
| + | *urease gene (or rather, the conglomeration of 6 genes that's required to make it). We're in the process of trying to biobrick urease, but it's not going very well. | ||
| + | *terminator BBa_B0015 | ||
| + | |||
| + | '''Device 3:''' | ||
| + | |||
| + | *ArsR1 promoter (with RBS included) - being made | ||
| + | *lambda cI coding gene - BBa_C0050 is available, but it looks pretty messy, we might want to look for another one or make our own. | ||
| + | *terminator BBa_B0015 | ||
| + | |||
| + | '''Device 4:''' | ||
| + | |||
| + | *ArsR2 promoter (with RBS included) - being made | ||
| + | *LacZ gene - BBa_E0033 is available, but we're in the process of making one that doesn't contain a multiple cloning site. | ||
| + | *Terminator BBa_B0015 | ||
| + | |||
| + | |||
| + | |||
| + | === Preliminary experiments === | ||
| + | |||
| + | * Playing with different copy numbers of plasmids and sequences of promoters to see if we can develop two arsenic sensitive promoters of different sensitivities | ||
| + | |||
| + | '''Without genetic modification:''' | ||
| + | * The pH of urea in solution | ||
| + | * The pH of lactose and urea in solution mixed together | ||
| + | * The change in pH when urea is digested by urease | ||
| + | * The change in pH when urea is digested by urease and lactose is present | ||
| + | * The change in pH when lactose is digested by lacZ when urea is present | ||
| + | |||
| + | '''With genetic modification:''' | ||
| + | |||
| + | We would hopefully have the lacZ system already from the simpler arsenic setup. So that's Device 3 sorted, but we need to engineer functioning Devices 1 and 2. | ||
| + | * Device 1: Sticking urease onto the end of the lac regulatory region. This shouldn't be a problem, as the regulatory region is available, and the whole thing is a reasonably simple procedure. We'd need to find out if it works, but hopefully... | ||
| + | * Device 2: This part is likely to be a nightmare. We'd have the arsenic sensor available from previous experiments, but we need to find a repressor that would completely neutralise the effects of the expressed urease. There's a number of ways of doing this, but it's likely to be kind of fiddly. It might be an idea to first engineer Device 1, then try it out by just putting in different repressor molecules manually into the sample to see which ones work best, and only then going on to genetically engineer the best one next to the ars sensor as detailed. | ||
| + | |||
| + | == Possible mechanism 3 - ultra ambitious == | ||
| + | |||
| + | This mechanism relies on the existence of a number of different ars operon style promoters with varying sensitivities to arsenic. I don't know if such things actually exist. If they don't, it's not actually worth it reading any further. However, if they do: | ||
| + | |||
| + | This mechanism is an elaboration on mechanism 2, combining pH as an indicator with fluorescence. | ||
| + | |||
| + | It also requires promoters that need 2 activator molecules to switch them on. As far as I know, such things exist, with the activator molecule binding to the specific gene sequence, and the co-activator binding to the activator and increasing the gene expression. The same coactivator can act on a number of different promoters, since it is only the activator part that's sequence specific. | ||
| + | |||
| + | Anyway. | ||
| + | |||
| + | We leave Device 1 as it is. | ||
| + | We leave Device 2 as it is, except that along with a repressor, it also expresses a coactivator molecule (lets call it CA1) | ||
| + | We leave Device 3 as it is. | ||
| + | |||
| + | And then we add another couple of devices: | ||
| + | |||
| + | '''Device 4:''' An ars operon that's activated at 10 ppb, that produces an activator molecule A1 | ||
| + | |||
| + | '''Device 5:''' promoter activated by A1+CA1, with a fluorescent gene attached, eg. GFP. | ||
| + | |||
| + | |||
| + | |||
| + | Now, '''if we have ars promoters that can bind two repressors''', we can really play around with this: | ||
| + | |||
| + | '''Device 6:''' an ars operon that's activated at 15 ppb, that produces activator molecule A2, as well as repressor molecule R1 which shuts down Device 4. | ||
| + | |||
| + | '''Device 7:''' promoter activated by A2+CA1, with a fluorescent gene attached, eg. YFP. | ||
| + | |||
| + | '''Device 8:''' activated at 20 ppb, produces A3 and R2, R2 shuts down Device 6 | ||
| + | |||
| + | '''Device 9:''' A3+CA1 make colour | ||
| + | |||
| + | etc. potentially ad infinitum, with each colour repressing the previous colour. | ||
| + | |||
| + | |||
| + | |||
| + | '''Failing that:''' | ||
| + | Without the promoters that bind 2 repressors, it's still possible to incorporate the colours, it's just that the number of colours we can incorporate will be limited. | ||
| + | We can do this by stopping at Device 5, then making Device 3 produce a different coactivator CA2, and having | ||
| + | |||
| + | '''Device 6:''' ars operon that's activated at, say, 30 ppb, produces A2 | ||
| + | |||
| + | '''Device 7:'''A2+CA2 produce a colour, eg. RFP | ||
| + | |||
| + | In this case, either another mechanism should be used to deactivate Device4+5, or Device 7 should produce a colour that's darker than that of Device 5, and thus still clearly visible even when both are active at the same time. | ||
| + | |||
| + | So what we get with the latter, simpler version is a display as follows: | ||
| + | |||
| + | Arsenic conc. (ppb) pH Colony colour | ||
| + | 0 9.5 none | ||
| + | 5 7 none | ||
| + | 10 7 green | ||
| + | 20 4.5 green | ||
| + | 30 4.5 red | ||
| + | |||
| + | The results for the more complex version above are pretty similar, except with a lot more different concentrations, and a proper rainbow spectrum of colours. | ||
| + | |||
| + | |||
| + | '''Advantage:''' once a colony of bacteria is engineered, no matter how complex the genetic engineering was, it's still very simple to grow. So provided this version was made in real life, it would still have all the advantages of the pH system we discussed - it would be cheap and easy to use in the field, only requiring a bit of lactose and indicator. However, this version would have the added advantage that it would also be possible to determine the concentration more precisely by consulting the above table if a fluorescence microscope was used. It could be a high throughput reasonably detailed technique used in labs as well as a quick easy test used in the field. A sort of all-in-one. Eureka. | ||
| Line 47: | Line 221: | ||
| - | == | + | == Model == |
| - | + | This is the latest modelling diagram of the arsenic biosensor | |
| + | [http://2006.igem.org/Arsenic_Sensor_Model Biosensor Model] | ||
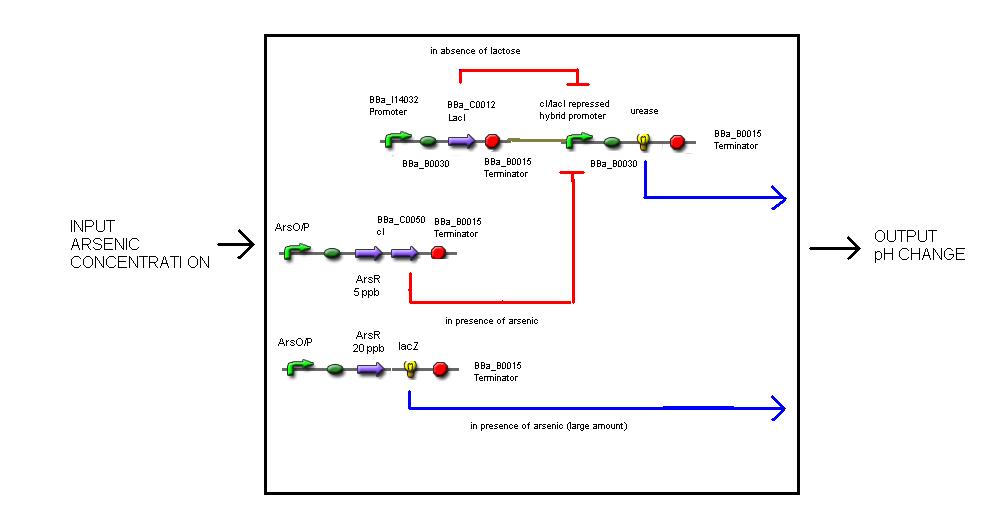
| - | + | The three levels of abstraction for the biosensor are :- | |
| - | + | [[Image: Systems_level_abstract.JPG|600px|thumb|center|Systems Level Diagram]] | |
| - | + | Systems level - the input is arsenic concentration and the output is pH change. | |
| - | + | ||
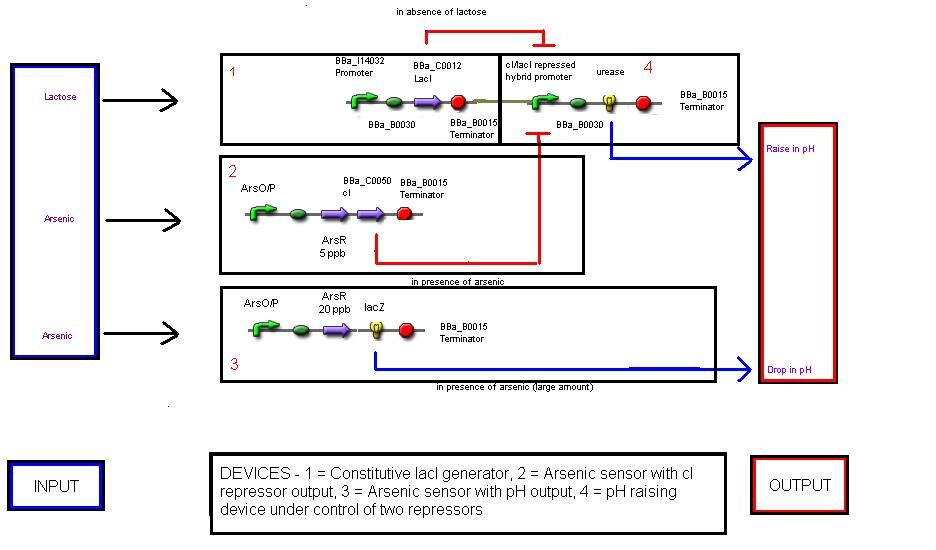
| + | [[Image: Device_level.JPG|600px|thumb|center|Device Level Diagram]] | ||
| + | Devices level - there are four devices which interact in the system | ||
| + | |||
| + | [[Image: Parts_level_abstract.JPG|600px|thumb|center|Parts Level Diagram]] | ||
| + | There are five parts taken from the registry, and six parts designed by our team | ||
| + | |||
| + | [http://2006.igem.org/University_of_Edinburgh_2006 Main page] | ||
| + | |||
| + | __NOTOC__ | ||
Latest revision as of 10:06, 18 October 2006
This is our first formalised project idea. The details are as follows:
Aim
Develop a bacterial biosensor that responds to a range of arsenic concentrations and produces a change in pH that can be calibrated in relation with the arsenic concentration.
The worst mass poisoning the world has ever known
It is estimated that at this very moment up to 100 million people across the world are being poisoned due to the presence of arsenic in their drinking water. One in a hundred of these people will develop a fatal cancer as a result, and countless others will get ill and suffer as the poison slowly damages their bodies.
This problem is greatest in one of the world’s poorest countries – Bangladesh. Because of the combination of regular yearly floods and poor sanitation, the available fresh water is swarming with harmful diseases. In an attempt to alleviate this problem, a large number of tube-wells were dug all across the country in order to provide people with clean water. However, in a cruel twist of fate, a lot of these wells were dug through a sedimentary layer containing arsenic.
Arsenic wasn't a standard thing to test for at the time, and by the time the first signs of the poisoning emerged, it was already too late – out of the estimated 10 million wells dug by now, at least 1 in 4, and possibly almost as many as a half of these wells contain water with harmful levels of arsenic. The organisations working in present day Bangladesh do carry out routine tests for arsenic in new wells, and arsenic mitigation efforts are being carried out, but the immense scale of the problem means these are basically just a drop in the ocean.
So the choice facing a poor rural family in Bangladesh is one between a quick death from disease if they drink the water from the rivers, or the potential of slow death from poisoning if they drink the water from the wells. The problem is that they don’t know which wells are poisoned, and because the effects of arsenic are slow and cumulative, by the time they find out, it’s already too late.
1 in 4 wells. Would you play Russian roulette with your children’s lives?
The project
Scientists around the world have been working to change this. The distribution of arsenic in the wells is quite irregular, so if a cheap, easy-to-use arsenic detector was to become available, all the wells could be tested, and safe ones could be found – something that is currently financially unfeasible because of the instruments and expertise required.
A lot of research has been done into arsenic biosensors already, and some have already been created, mostly using fluorescence. The components are available, there are gene promoters that respond to the presence of arsenic, and various different outputs are possible. The next stage is really more of an engineering question: how do you come up with an optimal design that’s easy to use in the field?
Our suggested project uses a pH change as a signal for the presence of arsenic, improving upon previous designs because the change is easily detectable using a pH meter, or even just a couple of drops of indicator. It is also potentially possible to make the system detect a large range of arsenic concentrations, because the change in pH is gradual.
Not only is such a device both practical and marketable, it is ethically imperative to produce it. Using the cutting edge of modern science, it really is possible to change the world.
Possible mechanism 1 - the bare minimum
Outline
INPUT [Arsenate/Arsenite] -> Detector -> Reporter -> OUTPUT [H+]
The arsR gene codes for a repressor which binds to the arsenic promoter in the absence of arsenate or arsenite. These two genes can be linked to the lacZ gene, in order to place lactose metabolism under the control of an arsenic activated promoter.
The ability to metabolise lactose should result in the acidification of the medium, and an output of H+ ions, giving a pH response to the input of arsenate/arsenite molecules.
It should be possible to obtain a curve for pH vs. arsenic concentration, where a range of pH values representing an unsafe level, a safe level, or an intermediate level of arsenic is present can be elucidated after calibrating the biosensor with known concentrations of arsenic.
Parts needed: Arsenic binding repressor protein Arsenic repressor protein responsive promoter lacZ protein coding gene
Preliminary experiments
- How fast (if at all) E. coli change the pH of the medium when the lacZ gene is activated.
- Playing with the lacZ gene attached to a promoter we can easily activate in the lab, so that we can see how the pH changes with regards to bacterial density, quantity of lactose present, growth stage the bacteria are in, etc.
- Testing the response of the reporter genes when ArsR is bound by arsenic
A lot of this project will be calibrating the sensor, finding the time to reach a steady state, finding out which amount of cells is ideal for reaching the steady state which gives distinct results for each pH range, and finding out which pH range represents which concentration of arsenic. It would be ideal to have the threshold for the legal limit of arsenic at the point of the curve which gives the most accurate result.
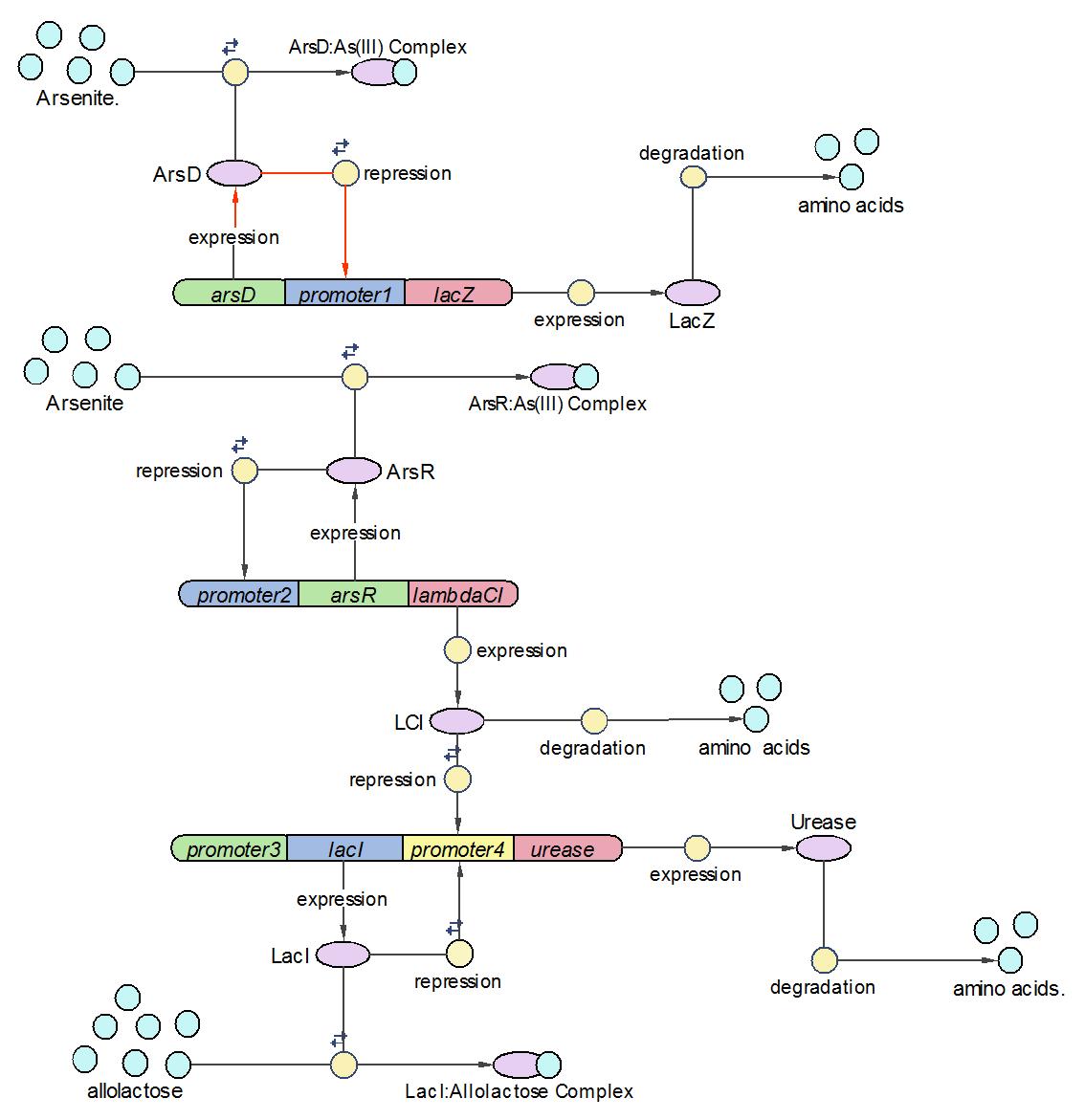
Possible mechanism 2 - more ambitious
Get the powerpoint animation on how the system works [http://2006.igem.org/Image:Presentation_only_1.ppt here!]
After discussion, it was decided that it would be unfeasible to try and engineer a sensitive enough analogue output just using the pH - instead, a set of a couple of defined 'stepped' outputs would be preferable.
In order to increase the range that is indicated by the biosensor, the following mechanism might be worth considering.
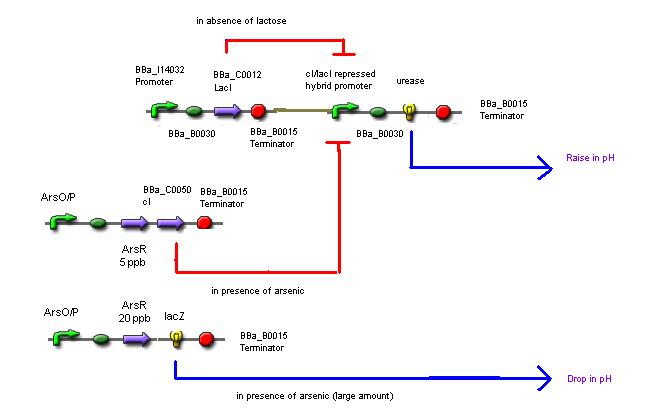
Sample: Water sample + Lactose + Urea + Bacterial culture
The mechanism involves 4 'devices' inside the bacteria.
Device 1: The lac operon regulatory region is connected to the LuxR coding gene.
Device 2: The LuxR/cI responsive promoter is connected to the gene for urease which, if activated, catalyses the conversion of urea into ammonia and carbon dioxide, raising the pH.
Device 3: A promoter that's regulated by a repressor very sensitive to arsenic. Thus anything above 5 ppb of arsenic would bind to the repressor and enable gene expression. This promoter is connected to a gene that expresses the lambda cI molecule, thus keeping the pH at neutral.
Device 4: A promoter regulated by a repressor that's somewhat less sensitive to arsenic, so only switches on at, say, 20 ppb. Connected to lacZ, as detailed above in Possible mechanism 1. The LacZ breaks down the lactose present, shutting down device 1.
So, if all goes according to plan, we would get the following results:
If no arsenic is present: The lac operon in Device 1 detects the presence of lactose that's been put in, activates the expression of the genes behind it, and urease catalyses the reaction that raises the pH of the surrounding solution. So the pH with no arsenic present is around 9-10.
If 5 ppb arsenic is present: The arsenic activates Device 2, which produces a repressor that deactivates Device 1. No enzyme is produced, and the pH remains neutral (or maybe slightly higher than neutral because urea is present, but there's still a distinguishable difference). 5ppb arsenic = pH 7
If 20 ppb arsenic is present: The arsenic activates Device 3, which produces lacZ, which digests the lactose in the solution and turns it acidic. Device 2 is still switched on and Device 1 is still switched off accordingly, so there are no problems with the acid and the alkali neutralising one another. 20ppb arsenic = pH 4.5
The figures for the concentrations and the pH are basically currently just made up, it would take weeks of calibration in the lab to tune everything properly and see what's feasible. Whether the E. coli survives the given range of pH or not is basically irrelevant, as each infusion of bacteria would be single use, and neither the alkali nor the acid will be produced until the bacteria are already sitting in the test sample.
Parts required
Device 1:
- RBS - BBa_B0030
- Promoter constitutively switched on. Part BBa_I14032, promoter P(Lac) IQ
- LacI gene: present in the registry, BBa_C0012. Has an LVA degradation tail attached, not sure if we need that. Doesn’t say it’s been found to be working, maybe we could make our own?
- Terminator: say BBa_B0015? It seems to be the most efficient one that works. We want it to be efficient, right?
- LacI regulated promoter: BBa_R0011, extensively tested and found to work. Mutated from a Lambda phage. Should be fine to use this one.
- LuxR coding gene - part BBa_C0062, seems like a good part, but there's no reports on whether it works.
- Terminator: BBa_B0015
Device 2:
- RBS - BBa_B0030
- Part BBa_R0065 - lambda cI and luxR regulated promoter
- urease gene (or rather, the conglomeration of 6 genes that's required to make it). We're in the process of trying to biobrick urease, but it's not going very well.
- terminator BBa_B0015
Device 3:
- ArsR1 promoter (with RBS included) - being made
- lambda cI coding gene - BBa_C0050 is available, but it looks pretty messy, we might want to look for another one or make our own.
- terminator BBa_B0015
Device 4:
- ArsR2 promoter (with RBS included) - being made
- LacZ gene - BBa_E0033 is available, but we're in the process of making one that doesn't contain a multiple cloning site.
- Terminator BBa_B0015
Preliminary experiments
- Playing with different copy numbers of plasmids and sequences of promoters to see if we can develop two arsenic sensitive promoters of different sensitivities
Without genetic modification:
- The pH of urea in solution
- The pH of lactose and urea in solution mixed together
- The change in pH when urea is digested by urease
- The change in pH when urea is digested by urease and lactose is present
- The change in pH when lactose is digested by lacZ when urea is present
With genetic modification:
We would hopefully have the lacZ system already from the simpler arsenic setup. So that's Device 3 sorted, but we need to engineer functioning Devices 1 and 2.
- Device 1: Sticking urease onto the end of the lac regulatory region. This shouldn't be a problem, as the regulatory region is available, and the whole thing is a reasonably simple procedure. We'd need to find out if it works, but hopefully...
- Device 2: This part is likely to be a nightmare. We'd have the arsenic sensor available from previous experiments, but we need to find a repressor that would completely neutralise the effects of the expressed urease. There's a number of ways of doing this, but it's likely to be kind of fiddly. It might be an idea to first engineer Device 1, then try it out by just putting in different repressor molecules manually into the sample to see which ones work best, and only then going on to genetically engineer the best one next to the ars sensor as detailed.
Possible mechanism 3 - ultra ambitious
This mechanism relies on the existence of a number of different ars operon style promoters with varying sensitivities to arsenic. I don't know if such things actually exist. If they don't, it's not actually worth it reading any further. However, if they do:
This mechanism is an elaboration on mechanism 2, combining pH as an indicator with fluorescence.
It also requires promoters that need 2 activator molecules to switch them on. As far as I know, such things exist, with the activator molecule binding to the specific gene sequence, and the co-activator binding to the activator and increasing the gene expression. The same coactivator can act on a number of different promoters, since it is only the activator part that's sequence specific.
Anyway.
We leave Device 1 as it is. We leave Device 2 as it is, except that along with a repressor, it also expresses a coactivator molecule (lets call it CA1) We leave Device 3 as it is.
And then we add another couple of devices:
Device 4: An ars operon that's activated at 10 ppb, that produces an activator molecule A1
Device 5: promoter activated by A1+CA1, with a fluorescent gene attached, eg. GFP.
Now, if we have ars promoters that can bind two repressors, we can really play around with this:
Device 6: an ars operon that's activated at 15 ppb, that produces activator molecule A2, as well as repressor molecule R1 which shuts down Device 4.
Device 7: promoter activated by A2+CA1, with a fluorescent gene attached, eg. YFP.
Device 8: activated at 20 ppb, produces A3 and R2, R2 shuts down Device 6
Device 9: A3+CA1 make colour
etc. potentially ad infinitum, with each colour repressing the previous colour.
Failing that: Without the promoters that bind 2 repressors, it's still possible to incorporate the colours, it's just that the number of colours we can incorporate will be limited. We can do this by stopping at Device 5, then making Device 3 produce a different coactivator CA2, and having
Device 6: ars operon that's activated at, say, 30 ppb, produces A2
Device 7:A2+CA2 produce a colour, eg. RFP
In this case, either another mechanism should be used to deactivate Device4+5, or Device 7 should produce a colour that's darker than that of Device 5, and thus still clearly visible even when both are active at the same time.
So what we get with the latter, simpler version is a display as follows:
Arsenic conc. (ppb) pH Colony colour
0 9.5 none
5 7 none
10 7 green
20 4.5 green
30 4.5 red
The results for the more complex version above are pretty similar, except with a lot more different concentrations, and a proper rainbow spectrum of colours.
Advantage: once a colony of bacteria is engineered, no matter how complex the genetic engineering was, it's still very simple to grow. So provided this version was made in real life, it would still have all the advantages of the pH system we discussed - it would be cheap and easy to use in the field, only requiring a bit of lactose and indicator. However, this version would have the added advantage that it would also be possible to determine the concentration more precisely by consulting the above table if a fluorescence microscope was used. It could be a high throughput reasonably detailed technique used in labs as well as a quick easy test used in the field. A sort of all-in-one. Eureka.
Problems
The arsenic promoter shows some background activity even when arsenic is not present. It may be possible to reduce this noise by adding an extra control method to repress the lacZ gene parallel to the action of the arsR product, with for example an antiswitch.
It will not be possible to get a linear relationship between pH and the arsenic concentration because the relationship between gene expression and the cofactor concentration is not linear.
The arsenic concentration has to be the limiting factor in the reaction to result in the steady state obtained being the correct pH in relation to arsenic concentration. If the cell number is the limiting factor the pH may not drop low enough to indicate the arsenic level present in the time taken before measuring.
Model
This is the latest modelling diagram of the arsenic biosensor
[http://2006.igem.org/Arsenic_Sensor_Model Biosensor Model]
The three levels of abstraction for the biosensor are :-
Systems level - the input is arsenic concentration and the output is pH change.
Devices level - there are four devices which interact in the system
There are five parts taken from the registry, and six parts designed by our team
[http://2006.igem.org/University_of_Edinburgh_2006 Main page]