University of California San Francisco 2006
From 2006.igem.org
(→Added Parts) |
|||
| (64 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
[[Image:Patrick_Angel.jpg|thumb|350px|''Guess what I put in your LB buffer!'']] | [[Image:Patrick_Angel.jpg|thumb|350px|''Guess what I put in your LB buffer!'']] | ||
== The University of California at San Francisco 2006 iGEM Team == | == The University of California at San Francisco 2006 iGEM Team == | ||
| - | The international Genetically Engineered Machine Competition is an opportunity for emerging young scientists of all disciplines to tinker with the proverbial Rube Goldberg machine of life | + | Mimicking remote controlled cars, we have developed a strain of bacteria that can switch the direction in which it swims based on an external signal. |
| - | [[Image: | + | |
| + | The genetic program required to perform this function involves reengineered sensors (the nose) and a genetic switch that is linked to the external input. | ||
| + | |||
| + | This project showcases a technology for the creation of multiple independent sensing/signaling pathways that can operate simultaneously in the cell. The ability to design signaling pathways de novo will have an impact on biotechnological applications, including biosensing, environmental niche detection, and cell-based therapeutics. | ||
| + | |||
| + | The international Genetically Engineered Machine Competition is an opportunity for emerging young scientists of all disciplines to tinker with the proverbial Rube Goldberg machine of life. | ||
| + | [[Image:logo2.jpg|thumb|200px|''It's this easy'']] | ||
| + | |||
== Team Members == | == Team Members == | ||
| - | + | *Patrick Visperas - [http://www.ucdavis.edu UC Davis] | |
| - | *Patrick Visperas | + | *Kevin Shay - Mission High School |
| - | *Kevin Shay | + | *Ala Trusina - [http://www.ucsf.edu UCSF] |
| - | *Matt Eames | + | *Matt Eames - [http://www.ucsf.edu UCSF] |
| + | |||
== Faculty Advisors == | == Faculty Advisors == | ||
| - | *[http://www.voigtlab.ucsf.edu/ Chris Voigt] | + | *[http://www.voigtlab.ucsf.edu/ Chris Voigt] - [http://www.ucsf.edu UCSF] |
| - | *[http://kortemmelab.ucsf.edu/ Tanja Kortemme] | + | *[http://kortemmelab.ucsf.edu/ Tanja Kortemme] - [http://www.ucsf.edu UCSF] |
| + | |||
== Aspirations, Dreams... == | == Aspirations, Dreams... == | ||
| - | [[Image: | + | [[Image:choice.jpg|thumb|left|250px|''Which chemoattractant would YOU choose?'']] |
| - | + | ''What if we could control chemotaxis? Could we put bacteria on plate and steer them through a maze? Would it be like operating a remote-controlled car?'' | |
| - | *'''Design and construction of the orthogonal interaction:''' An existing orthogonal interaction, to be used as a starting point prior to computational redesign, was discovered by [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1860813&query_hl=2&itool=pubmed_docsum Liu and Parkinson] in 1991. | + | |
| - | + | These were the questions that inspired team UCSF to tinker with bacterial chemotaxis. Naturally, bacteria don't have the same electric components that your remote-controlled buggy does, but there are parts we can harness to control motility. ''Escherichia coli'', for example, already possess five different chemoreceptors which respond to different attractants and repellants. If we were to place the bugs in between two separate chemicals, we might be able to control the direction of swimming by manipulating which receptor's signal we interpret. | |
| - | *'''Implementation of the genetic switch:''' This will allow us to toggle between the selective expression of either | + | |
| + | The UCSF iGEM team has endeavored to control chemotaxis by selectively expressing ''CheW'' mutants which will participate in an orthogonal interaction with the methyl-accepting chemotaxis protein Tar. The project will be broken down into three major components: | ||
| + | *'''Design and construction of the orthogonal interaction:''' An existing orthogonal interaction found in the ''Tsr'' receptor, to be used as a starting point prior to computational redesign, was discovered by [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1860813&query_hl=2&itool=pubmed_docsum Liu and Parkinson] in 1991. | ||
| + | *'''Implementation of the genetic switch:''' This will allow us to toggle between the selective expression of either ''CheW''. | ||
*'''Validation of chemotactic properties through specially developed assay:''' originally developed by the good folks at the [http://www.physics.upenn.edu/facultyinfo/goulian.html Goulian Lab] in 2006. | *'''Validation of chemotactic properties through specially developed assay:''' originally developed by the good folks at the [http://www.physics.upenn.edu/facultyinfo/goulian.html Goulian Lab] in 2006. | ||
| - | [[Image: | + | |
| - | + | == The Orthogonal Interaction and Receptor Specificity == | |
| + | To selectively discriminate between the chemoreceptors, we will re-engineer the signaling protein ''CheW'' (which does not differentiate between receptors in its native state), to recognize only one receptor. This will require the development of an orthogonal interaction - that is two interacting partners that have no cross-talk between orhtogonal partners. | ||
| + | [[Image:cartoon4.jpg|thumb|left|200px|''An orthogonal interaction for all seasons'']] | ||
| + | |||
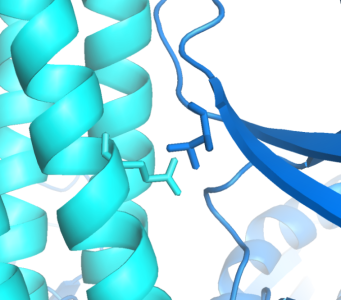
| + | Implementing an orthogonal interaction typicaly requires detailed knowledge of the interface. While no structure for the ''CheW-Tar'' complex yet exists, their unbound structures have been characterized. By computationally docking them together, we can observe that residues identified in the ''Tsr'' receptor as to having destabilizing/recovery properties for binding do indeed make contact in complex. This interface can be further redesigned using computational approaches. Such methods have the ability to generate infinite signal capacity. | ||
| + | [[Image:complex1.jpg|thumb|right|200px|''Docked and loaded: Mutate these two residues and you get orthogonal bonding activity'']] | ||
| + | |||
| + | Now we also need two receptors which are responsive to different attractants. | ||
| + | |||
Much work is available in the literature regarding the changing of chemoreceptor specificity. Some of the options include: | Much work is available in the literature regarding the changing of chemoreceptor specificity. Some of the options include: | ||
*Directed evolution | *Directed evolution | ||
*Computational redesign | *Computational redesign | ||
| - | *Chimeric construction (perisplasmic and cytoplasmic domains of the different receptors can be swapped) | + | *Chimeric construction (perisplasmic and cytoplasmic domains of the different receptors can be swapped) |
| - | We will be using a mutant version of the E. coli | + | |
| - | == | + | We will be using a mutant version of the ''E. coli Tar'' (asparate-sensitive) receptor that has been evolved to respond to phenylalanine, a natural chemorepellant. The responsiveness of this mutant has been characterized and was found to have comparable chemotactic capabilities to that of wild-type ''Tar''. Our phenylalanine-responsive receptor will bind to wild-type ''CheW'', and our aspartate-responsive ''Tar'' will have its cytoplasmic domain mutated to accommodate the mutated ''CheW'' as its binding partner. |
| - | To eliminate interference from other naturally occuring receptors in E. coli, a strain with all receptors (''Tar, Tsr, Tap, Trg, Aer'') knocked out, provided by the Parkinson Lab, will be used in this work. | + | |
| - | == | + | '''Mutated Cytoplasmic Domain of ''Tar'' for orthogonal interaction''' |
| - | [[Image: | + | *EA400 |
| + | '''Mutated ''CheW'' for orthogonal interaction''' | ||
| + | *VM108 | ||
| + | '''Mutated ''Tar'' residues to generate phenylalanine response''' | ||
| + | *IA2, LI21, SC27, SP32, VE41, SC43, RW73, MV76, AS101, YF149, VA196, DV249, ST255 | ||
| + | |||
| + | == The Genetic Switch == | ||
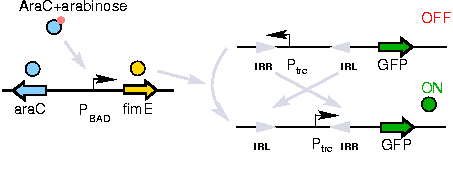
| + | A previously developed genetic switch using ''fimE'' to invert a nucleotide sequence (including a promoter) will be used to selectively control expression of the ''CheW'' pairs. | ||
| + | [[Image:switch.jpg|thumb|right|250px|''Light goes on, light goes off'']] | ||
| + | |||
| + | The activation of the switching mechanism is under the control of arabinose. The genetic inversion is irreversible. | ||
| + | |||
| + | == Chassis == | ||
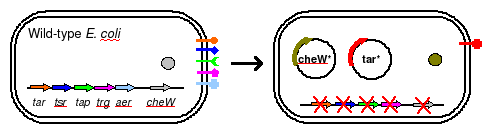
| + | [[Image:strain1.jpg|thumb|left|250px|''Break it down to build it up'']] | ||
| + | To eliminate interference from other naturally occuring receptors in ''E. coli'', a strain with all receptors (''Tar, Tsr, Tap, Trg, Aer'') knocked out, provided by the Parkinson Lab, will be used in this work. The UCSF iGEM deleted ''CheW'' from this strain so that all ''CheW'' might be expression off the plasmid. | ||
| + | |||
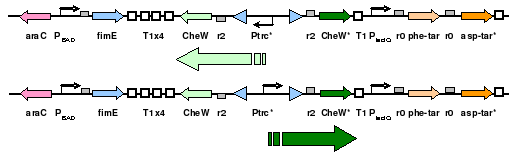
| + | The plasmid will posses both constituitively expressed ''Tar'' varients, and will incorporate selective ''CheW'' control by means of a genetic switch. The use of redundant pairs has required condon randomization to prevent unwanted recombination events. | ||
| + | [[Image:chassis.jpg|thumb|300px|''A two-state system'']] | ||
| + | |||
| + | == Assaying Chemotaxis == | ||
| + | We will use an assay developed by the Goulian Lab, which works almost as simply as spotting a bacterial culture on a plate with an attractant gradient. | ||
| + | |||
| + | The use of minimal media will generate a stronger phenotype. We established the gradient by linearly spotting the desired chemoattractant down the plate and letting is diffuse over several hours. The eventual gradient will have a Gaussian profile. | ||
| + | |||
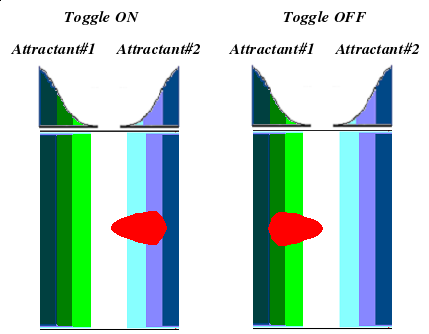
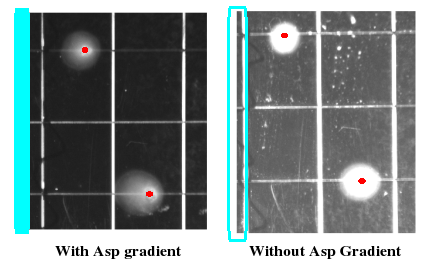
| + | We can observe chemotactic behavior one day after spotting bacterial culture in the gradient. While cultures spotted on plates without attractant will spread uniformly outward, those spotted within the gradient will migrate in a highly elliptical pattern, biased towards higher concentrations. | ||
| + | [[Image:assay.jpg|thumb|right|250px|''A sneak peak at some sweet results'']] | ||
| + | |||
| + | == Added Parts == | ||
| + | For the consideration of the judges, the parts: | ||
| + | |||
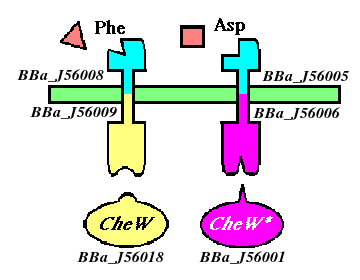
| + | '''Ligand domains, sensors''' | ||
| + | *[http://partsregistry.org/Part:BBa_J56005 BBa_J56005]: ''Tar_lig'', aspartate-sensitive ligand-binding domain of chemoreceptor | ||
| + | *[http://partsregistry.org/Part:BBa_J56008 BBa_J56008]: ''PheTar_lig'', phenylalanine-sensitive ligand-binding domain of chemoreceptor | ||
| + | '''Signaling domains, sensors''' | ||
| + | *[http://partsregistry.org/Part:BBa_J56006 BBa_J56006]: ''Tar_sig'', signaling domain of Tar, mutated to bind with BBa_J56001 | ||
| + | *[http://partsregistry.org/Part:BBa_J56009 BBa_J56009]: ''PheTar_sig'', signaling domain of PheTar, binds to w.t. ''CheW'' | ||
| + | '''Complete sensors''' | ||
| + | *[http://partsregistry.org/Part:BBa_J56002 BBa_J56004]: ''Tar'', aspartate receptor mutated to bind with BBa_J56001 | ||
| + | *[http://partsregistry.org/Part:BBa_J56003 BBa_J56007]: ''PheTar'', phenylalanine receptor mutated to respond to phenylalanine | ||
| + | '''Signaling proteins''' | ||
| + | *[http://partsregistry.org/Part:BBa_J56001 BBa_J56001]: ''CheW_mut'', mutated to bind with orthogonal partner BBa_J56004 | ||
| + | *[http://partsregistry.org/Part:BBa_J56018 BBa_J56018]: ''CheW'', wild-type CheW | ||
| + | |||
| + | For the considerattion of the judges, the system | ||
| + | *[http://partsregistry.org/Part:BBa_J56017 BBa_J56017]: Composite sequence, includes orthogonal ''CheW''-receptor pairs and the genetic switch | ||
| + | |||
== Thanks == | == Thanks == | ||
| + | *[http://www.chemeng.uiuc.edu/Faculty/rao.html Chris Rao] | ||
*[http://www.biology.utah.edu/faculty2.php?inum=6 Parkinson Lab] | *[http://www.biology.utah.edu/faculty2.php?inum=6 Parkinson Lab] | ||
*[http://www.physics.upenn.edu/facultyinfo/goulian.html Goulian Lab] | *[http://www.physics.upenn.edu/facultyinfo/goulian.html Goulian Lab] | ||
| + | |||
== Links == | == Links == | ||
*[http://www.ucsf.edu UCSF] | *[http://www.ucsf.edu UCSF] | ||
Latest revision as of 19:02, 31 October 2006
Contents |
The University of California at San Francisco 2006 iGEM Team
Mimicking remote controlled cars, we have developed a strain of bacteria that can switch the direction in which it swims based on an external signal.
The genetic program required to perform this function involves reengineered sensors (the nose) and a genetic switch that is linked to the external input.
This project showcases a technology for the creation of multiple independent sensing/signaling pathways that can operate simultaneously in the cell. The ability to design signaling pathways de novo will have an impact on biotechnological applications, including biosensing, environmental niche detection, and cell-based therapeutics.
The international Genetically Engineered Machine Competition is an opportunity for emerging young scientists of all disciplines to tinker with the proverbial Rube Goldberg machine of life.
Team Members
- Patrick Visperas - [http://www.ucdavis.edu UC Davis]
- Kevin Shay - Mission High School
- Ala Trusina - [http://www.ucsf.edu UCSF]
- Matt Eames - [http://www.ucsf.edu UCSF]
Faculty Advisors
- [http://www.voigtlab.ucsf.edu/ Chris Voigt] - [http://www.ucsf.edu UCSF]
- [http://kortemmelab.ucsf.edu/ Tanja Kortemme] - [http://www.ucsf.edu UCSF]
Aspirations, Dreams...
What if we could control chemotaxis? Could we put bacteria on plate and steer them through a maze? Would it be like operating a remote-controlled car?
These were the questions that inspired team UCSF to tinker with bacterial chemotaxis. Naturally, bacteria don't have the same electric components that your remote-controlled buggy does, but there are parts we can harness to control motility. Escherichia coli, for example, already possess five different chemoreceptors which respond to different attractants and repellants. If we were to place the bugs in between two separate chemicals, we might be able to control the direction of swimming by manipulating which receptor's signal we interpret.
The UCSF iGEM team has endeavored to control chemotaxis by selectively expressing CheW mutants which will participate in an orthogonal interaction with the methyl-accepting chemotaxis protein Tar. The project will be broken down into three major components:
- Design and construction of the orthogonal interaction: An existing orthogonal interaction found in the Tsr receptor, to be used as a starting point prior to computational redesign, was discovered by [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1860813&query_hl=2&itool=pubmed_docsum Liu and Parkinson] in 1991.
- Implementation of the genetic switch: This will allow us to toggle between the selective expression of either CheW.
- Validation of chemotactic properties through specially developed assay: originally developed by the good folks at the [http://www.physics.upenn.edu/facultyinfo/goulian.html Goulian Lab] in 2006.
The Orthogonal Interaction and Receptor Specificity
To selectively discriminate between the chemoreceptors, we will re-engineer the signaling protein CheW (which does not differentiate between receptors in its native state), to recognize only one receptor. This will require the development of an orthogonal interaction - that is two interacting partners that have no cross-talk between orhtogonal partners.
Implementing an orthogonal interaction typicaly requires detailed knowledge of the interface. While no structure for the CheW-Tar complex yet exists, their unbound structures have been characterized. By computationally docking them together, we can observe that residues identified in the Tsr receptor as to having destabilizing/recovery properties for binding do indeed make contact in complex. This interface can be further redesigned using computational approaches. Such methods have the ability to generate infinite signal capacity.
Now we also need two receptors which are responsive to different attractants.
Much work is available in the literature regarding the changing of chemoreceptor specificity. Some of the options include:
- Directed evolution
- Computational redesign
- Chimeric construction (perisplasmic and cytoplasmic domains of the different receptors can be swapped)
We will be using a mutant version of the E. coli Tar (asparate-sensitive) receptor that has been evolved to respond to phenylalanine, a natural chemorepellant. The responsiveness of this mutant has been characterized and was found to have comparable chemotactic capabilities to that of wild-type Tar. Our phenylalanine-responsive receptor will bind to wild-type CheW, and our aspartate-responsive Tar will have its cytoplasmic domain mutated to accommodate the mutated CheW as its binding partner.
Mutated Cytoplasmic Domain of Tar for orthogonal interaction
- EA400
Mutated CheW for orthogonal interaction
- VM108
Mutated Tar residues to generate phenylalanine response
- IA2, LI21, SC27, SP32, VE41, SC43, RW73, MV76, AS101, YF149, VA196, DV249, ST255
The Genetic Switch
A previously developed genetic switch using fimE to invert a nucleotide sequence (including a promoter) will be used to selectively control expression of the CheW pairs.
The activation of the switching mechanism is under the control of arabinose. The genetic inversion is irreversible.
Chassis
To eliminate interference from other naturally occuring receptors in E. coli, a strain with all receptors (Tar, Tsr, Tap, Trg, Aer) knocked out, provided by the Parkinson Lab, will be used in this work. The UCSF iGEM deleted CheW from this strain so that all CheW might be expression off the plasmid.
The plasmid will posses both constituitively expressed Tar varients, and will incorporate selective CheW control by means of a genetic switch. The use of redundant pairs has required condon randomization to prevent unwanted recombination events.
Assaying Chemotaxis
We will use an assay developed by the Goulian Lab, which works almost as simply as spotting a bacterial culture on a plate with an attractant gradient.
The use of minimal media will generate a stronger phenotype. We established the gradient by linearly spotting the desired chemoattractant down the plate and letting is diffuse over several hours. The eventual gradient will have a Gaussian profile.
We can observe chemotactic behavior one day after spotting bacterial culture in the gradient. While cultures spotted on plates without attractant will spread uniformly outward, those spotted within the gradient will migrate in a highly elliptical pattern, biased towards higher concentrations.
Added Parts
For the consideration of the judges, the parts:
Ligand domains, sensors
- [http://partsregistry.org/Part:BBa_J56005 BBa_J56005]: Tar_lig, aspartate-sensitive ligand-binding domain of chemoreceptor
- [http://partsregistry.org/Part:BBa_J56008 BBa_J56008]: PheTar_lig, phenylalanine-sensitive ligand-binding domain of chemoreceptor
Signaling domains, sensors
- [http://partsregistry.org/Part:BBa_J56006 BBa_J56006]: Tar_sig, signaling domain of Tar, mutated to bind with BBa_J56001
- [http://partsregistry.org/Part:BBa_J56009 BBa_J56009]: PheTar_sig, signaling domain of PheTar, binds to w.t. CheW
Complete sensors
- [http://partsregistry.org/Part:BBa_J56002 BBa_J56004]: Tar, aspartate receptor mutated to bind with BBa_J56001
- [http://partsregistry.org/Part:BBa_J56003 BBa_J56007]: PheTar, phenylalanine receptor mutated to respond to phenylalanine
Signaling proteins
- [http://partsregistry.org/Part:BBa_J56001 BBa_J56001]: CheW_mut, mutated to bind with orthogonal partner BBa_J56004
- [http://partsregistry.org/Part:BBa_J56018 BBa_J56018]: CheW, wild-type CheW
For the considerattion of the judges, the system
- [http://partsregistry.org/Part:BBa_J56017 BBa_J56017]: Composite sequence, includes orthogonal CheW-receptor pairs and the genetic switch
Thanks
- [http://www.chemeng.uiuc.edu/Faculty/rao.html Chris Rao]
- [http://www.biology.utah.edu/faculty2.php?inum=6 Parkinson Lab]
- [http://www.physics.upenn.edu/facultyinfo/goulian.html Goulian Lab]
Links
- [http://www.ucsf.edu UCSF]