ETH2006 light
From 2006.igem.org
(→implementation alternatives) |
(→Test results: conceal strange facts) |
||
| (7 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | →[[ETH Zurich 2006#System modeling|back to main page]] | |
| - | + | == Light sensing device == | |
| - | === | + | The light sensing device's PoPS output activity should be a monotonic function of the light intensity the cell is subjected to. |
| + | |||
| + | === Implementation alternatives === | ||
An already existing light sensing device made by the coliroid project was reused: [http://www.nature.com/nature/journal/v438/n7067/abs/nature04405.html Synthetic biology: Engineering Escherichia coli to see light] (Nature 2005), [http://partsregistry.org/Featured_Parts:Light_Sensor light sensor device]. | An already existing light sensing device made by the coliroid project was reused: [http://www.nature.com/nature/journal/v438/n7067/abs/nature04405.html Synthetic biology: Engineering Escherichia coli to see light] (Nature 2005), [http://partsregistry.org/Featured_Parts:Light_Sensor light sensor device]. | ||
| - | |||
| - | |||
| - | |||
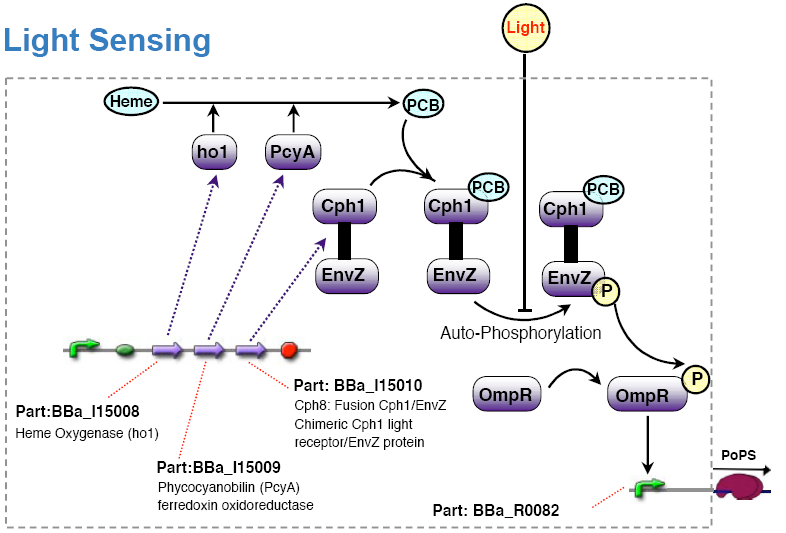
[[Image:ETH light sensing parts.png|600px|center]] | [[Image:ETH light sensing parts.png|600px|center]] | ||
| - | === | + | '''Note''': This light sensing device is ''low active'', that is no light results in high PoPS output, exposure to light in low PoPS. Technically, we would have to add an additional inverter to the system to comply with the specification. For simplicity, we decided to ''redefine'' the light input in such a way that no light means active. For experiments, however, it is anyway preferable if light means no input signal and thus no action. One can then operate under normal daylight conditions without stimulating the system inputs. |
| + | |||
| + | === Assembly procedure === | ||
Olga got the strain '''RU1012(KmR)''' and the plasmids '''pCPH8(CmR)''' and '''pPL-PCB(AmpR)''' from Christopher Voigt <cavoigt at picasso.ucsf.edu> [Engineering Escherichia Coli to see light; Nature vol. 438.]. | Olga got the strain '''RU1012(KmR)''' and the plasmids '''pCPH8(CmR)''' and '''pPL-PCB(AmpR)''' from Christopher Voigt <cavoigt at picasso.ucsf.edu> [Engineering Escherichia Coli to see light; Nature vol. 438.]. | ||
| Line 35: | Line 36: | ||
## run tests | ## run tests | ||
| - | === | + | === Test procedure === |
# plate saturated liquid culture out on LB+envZ(15mg/50ml) plates. | # plate saturated liquid culture out on LB+envZ(15mg/50ml) plates. | ||
| Line 41: | Line 42: | ||
# the colonies on the wrapped plate should remain colorless, the ones exposed to light should become dark. | # the colonies on the wrapped plate should remain colorless, the ones exposed to light should become dark. | ||
| - | In the auxiliaries to the nature publication they state regarding theyer light source: ".1331 W/cm^2 from 620-680nm (active state of the phytochrome) and .0132 W/cm^2 from 715-755nm (inactive state of the phytochrome)". 715-755nm is near infrared, 620-680nm is red. The best matching commonly available light sources I can find would be | + | In the auxiliaries to the nature publication they state regarding theyer light source: ".1331 W/cm^2 from 620-680nm (active state of the phytochrome) and .0132 W/cm^2 from 715-755nm (inactive state of the phytochrome)". 715-755nm is near infrared, 620-680nm is red. The best matching commonly available light sources I can find would be [http://en.wikipedia.org/wiki/Image:Red-YellowGreen-Blue_LED_spectra.gif| red LED] [http://en.wikipedia.org/wiki/Image:Spectra-Philips_32T8_natural_sunshine_fluorescent_light.jpg| white fluorescent light] and [http://en.wikipedia.org/wiki/Image:Spectrum_of_blue_sky.gif| indirect skylight]. A simple battery driven LED light source such as a bicycle backlight should work and could be put into the incubator, but would not be very powerful. A compact fluorescent light might be more appropriate but harder to get. |
| - | === | + | === Assembly progress === |
;2006-10-16: | ;2006-10-16: | ||
| Line 66: | Line 67: | ||
;2006-10-23: | ;2006-10-23: | ||
# mounted the plates with bacteria-side towards the light source to a white fluorescent lamp for approx 6h --[[User:Ajk|Ajk]] | # mounted the plates with bacteria-side towards the light source to a white fluorescent lamp for approx 6h --[[User:Ajk|Ajk]] | ||
| + | ;2006-10-29: | ||
| + | # made glycerol stocks of RU1012+pPCB+pCPH8, stored them in -80°C freezer at Sven Panke's lab --[[User:Ajk|Ajk]] | ||
| - | === | + | === Test results === |
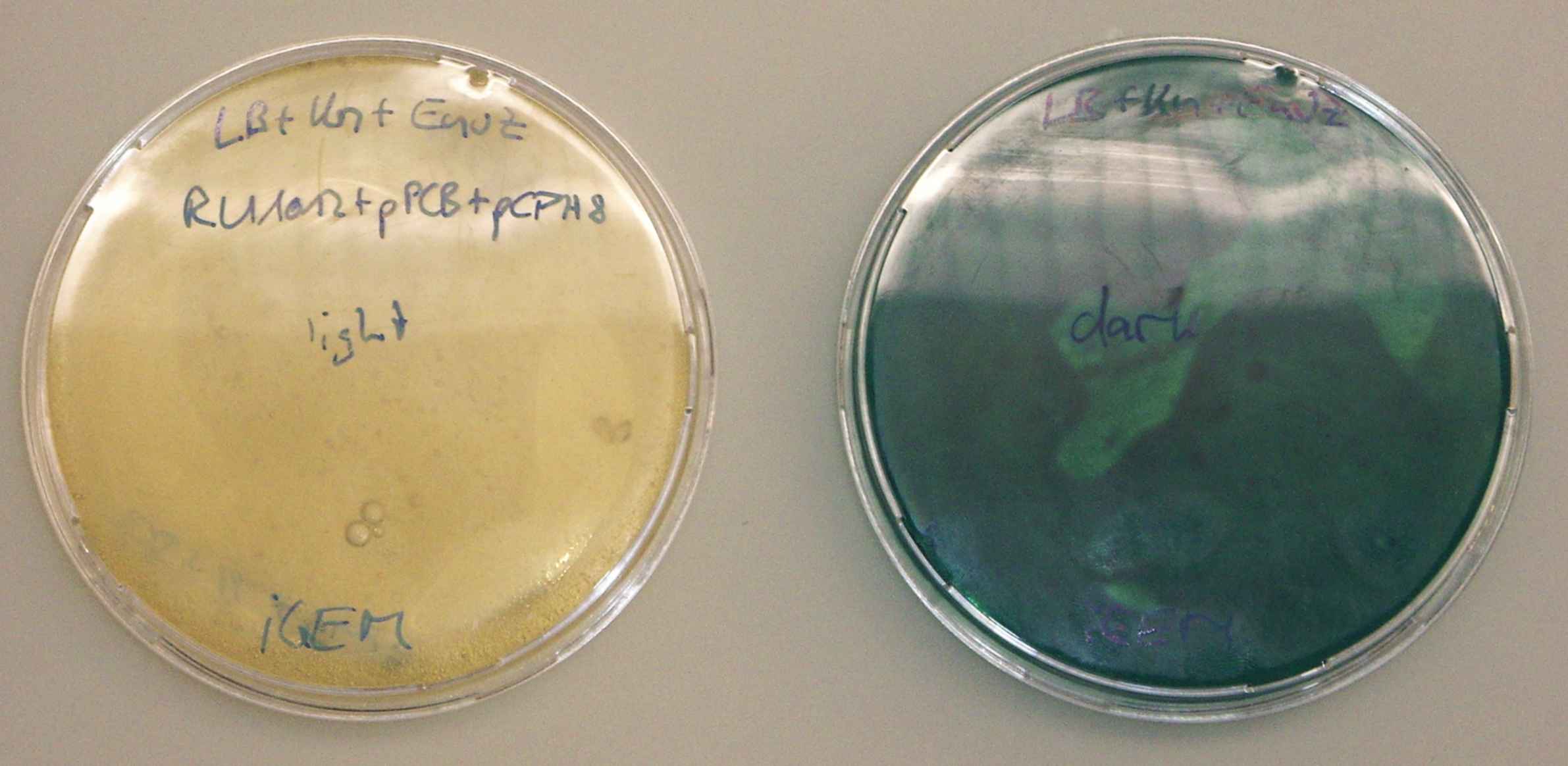
| - | The unwrapped plate stayed clear throughout the tests. The wrapped plate was deeply blue when it was inspected at the end | + | The unwrapped plate stayed clear throughout the tests. The wrapped plate was deeply blue when it was inspected at the end. --[[User:Ajk|Ajk]] 11:29, 23 October 2006 (EDT) |
[[Image:ETH06 Light sensing twoplatestest.jpg|600px|center]] | [[Image:ETH06 Light sensing twoplatestest.jpg|600px|center]] | ||
| - | === | + | === Parts === |
[http://partsregistry.org/Featured_Parts:Light_Sensor light sensor device#BioBricks parts list] | [http://partsregistry.org/Featured_Parts:Light_Sensor light sensor device#BioBricks parts list] | ||
| - | + | →[[ETH Zurich 2006#System modeling|back to main page]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | →[[ETH Zurich 2006# | + | |
Latest revision as of 18:11, 31 October 2006
Contents |
Light sensing device
The light sensing device's PoPS output activity should be a monotonic function of the light intensity the cell is subjected to.
Implementation alternatives
An already existing light sensing device made by the coliroid project was reused: [http://www.nature.com/nature/journal/v438/n7067/abs/nature04405.html Synthetic biology: Engineering Escherichia coli to see light] (Nature 2005), [http://partsregistry.org/Featured_Parts:Light_Sensor light sensor device].
Note: This light sensing device is low active, that is no light results in high PoPS output, exposure to light in low PoPS. Technically, we would have to add an additional inverter to the system to comply with the specification. For simplicity, we decided to redefine the light input in such a way that no light means active. For experiments, however, it is anyway preferable if light means no input signal and thus no action. One can then operate under normal daylight conditions without stimulating the system inputs.
Assembly procedure
Olga got the strain RU1012(KmR) and the plasmids pCPH8(CmR) and pPL-PCB(AmpR) from Christopher Voigt <cavoigt at picasso.ucsf.edu> [Engineering Escherichia Coli to see light; Nature vol. 438.].
A autonomous light sensing system can be obtained by cloning those two plasmids into the strain.
- plate RU1012 on LB+Km
- pick RU1012 cultures -> 5ml liquid culture (LB+Km)
-
- inoculate 200ml liquid culture
- grow for 3h
- wash twice (that means centrifuge, pipet off the liquid, dissolve in fresh LB?)
- make them competent
- make samples
- transform with both pCPH8 and pPL-PCB
- plate on
- LB+Cm
- LB+Ap
- LB+Ap+Cm
-
- if cultures survive on the Ap+Cm plate inoculate liquid culture 5ml LB+Km
- run tests
Test procedure
- plate saturated liquid culture out on LB+envZ(15mg/50ml) plates.
- wrap one plate in something opaque, expose both plates to light at 37°C.
- the colonies on the wrapped plate should remain colorless, the ones exposed to light should become dark.
In the auxiliaries to the nature publication they state regarding theyer light source: ".1331 W/cm^2 from 620-680nm (active state of the phytochrome) and .0132 W/cm^2 from 715-755nm (inactive state of the phytochrome)". 715-755nm is near infrared, 620-680nm is red. The best matching commonly available light sources I can find would be [http://en.wikipedia.org/wiki/Image:Red-YellowGreen-Blue_LED_spectra.gif| red LED] [http://en.wikipedia.org/wiki/Image:Spectra-Philips_32T8_natural_sunshine_fluorescent_light.jpg| white fluorescent light] and [http://en.wikipedia.org/wiki/Image:Spectrum_of_blue_sky.gif| indirect skylight]. A simple battery driven LED light source such as a bicycle backlight should work and could be put into the incubator, but would not be very powerful. A compact fluorescent light might be more appropriate but harder to get.
Assembly progress
- 2006-10-16
- 2006-10-17
- picked a RU1012 colony and inoculated a 5ml liquid culture (LB+Km), plate in fridge --Ajk
- 2006-10-18
- diluted the RU1012 liquid culture to 200ml and incubated it again --Ajk
- it turned out that my interpretation of the lab procedure notes I got was too minimalistic, had to set up a new 3ml liquid LB culture from another colony on the plate made th 16th. --Ajk
- 2006-10-19
- diluted yesterdays liquid culture to 2x50ml and grew it up to OD600=~.4 --Ajk
- poured plates 2x Amp and 4x Cm+Amp --Ajk
- washed the cultures twice with water, once with 10% glycerol --Ajk
- one shot of electroporesis with both pPCB and pCPH8 and one control without plasmids, leftover RU1012 in -80°C --Ajk
- plated on 4x Cm+Amp, 2x Cm, 2x Amp, control: 1x Cm, 1x Km leftover transformed cells in fridge --Ajk
- 2006-10-20
- found colonies on all plates except the Cm control which is perfect!! picked one of the colonies from a Cm+Amp plate into a 5ml liquid LB+Km culture for testing. --Ajk
- poured 2x LB+Km+envZ plates --Ajk
- 2006-10-21;
- plated 100ul liquid culture onto each of the LB+Km+envZ plates, wrapped one of them in tin foil and incubated both --Ajk
- 2006-10-23
- mounted the plates with bacteria-side towards the light source to a white fluorescent lamp for approx 6h --Ajk
- 2006-10-29
- made glycerol stocks of RU1012+pPCB+pCPH8, stored them in -80°C freezer at Sven Panke's lab --Ajk
Test results
The unwrapped plate stayed clear throughout the tests. The wrapped plate was deeply blue when it was inspected at the end. --Ajk 11:29, 23 October 2006 (EDT)
Parts
[http://partsregistry.org/Featured_Parts:Light_Sensor light sensor device#BioBricks parts list]