|
|
| (13 intermediate revisions not shown) |
| Line 64: |
Line 64: |
| | | | |
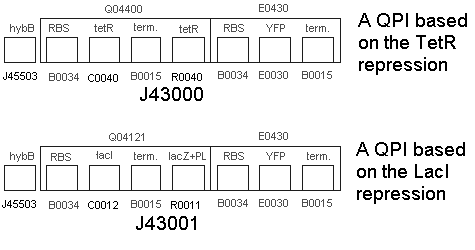
| | * We built the following two constructs:<br> | | * We built the following two constructs:<br> |
| - | :[[Image:untitled4.gif]] | + | :[[Image:untitled6.gif]] |
| | | | |
| | |} | | |} |
| Line 73: |
Line 73: |
| | <h3>Results</h3> | | <h3>Results</h3> |
| | | | |
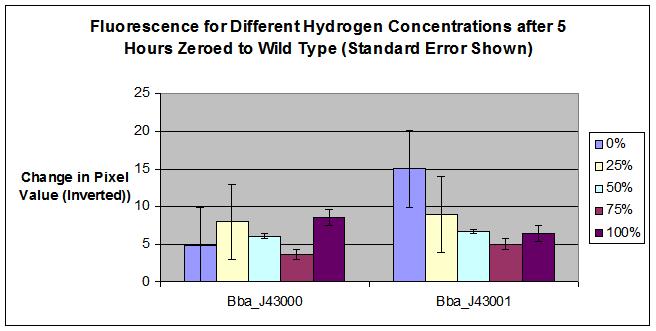
| - | :[[Image:results.jpeg]] | + | :[[Image:MSUResultsErrorbars.JPG]] |
| | + | |
| | + | * Part BBa_J43000 |
| | + | |
| | + | * No Hydrogen, No tetR = fluorescence |
| | + | * No Hydrogen, with tetR = Repressed Transcription at R0040 tetR, no fluorescence (except for some leakage). |
| | + | * Hydrogen + tetR = start trqanscription at hybB, produce tetR, increased repression of operon at R0040 tetR. no fluorescence. |
| | + | |
| | + | |
| | + | * Part BBa_J43001 |
| | + | |
| | + | * No Hydrogen, No inducer = no fluorescence (except for a bit of leakage possibly). |
| | + | * No Hydrogen, Inducer = Start Transcription at lacZ+PL, fluoresce. |
| | + | * Hydrogen + Inducer = start transcription at hybB, produce lacI, competitive inhibition of operon at lacZ+PL. Less fluorescence transcribed. |
| | + | |
| | |} | | |} |
| | | | |
| Line 81: |
Line 95: |
| | <h3>Discussion</h3> | | <h3>Discussion</h3> |
| | | | |
| - | * After three rounds of testing, we are unable to verify our hybB promoter as a hydrogen sensing promoter. However, the results do indicate that hydrogen sensing capabilities are probable | + | * Complications: |
| - | * The results from the test with J43000 were not at all expected. There are several points in the data that are unexplainable. It would appear as though some human error in testing or our tetR switch led to the inconsistencies. The tetR switch was reported to be an inverter in the registry. Therefore, these results could be explainable by a mistake in the registry. On the other hand, our results could be explained by our promoter functioning as an inverter as well. In that case, there should increasing fluorescence with hydrogen concentration.
| + | |
| - | * The results from the test with J43001 were also not as expected. The LacI switch was not known to be an inverter. However, our results show that the entire part was producing an inverted signal. Either the LacI switch is an inverter or the promoter itself could be an inverter. If either of these are true, we would see decreasing fluorescence with hydrogen concentration. | + | * BBa_J43000 was a failed part, we learned during completion of the construction and testing that there was a flaw in the design of the composite part. Although the hybB promoter does appear to detect hydrogen, the YFP is inhibited with the addition of tetR. Therefore, we cannot detect a change in fluorescence when hydrogen is added. |
| - | * Based on the tests, it would appear as though our promoter displays some inverting characteristics. This is the only possible outcome that could explain the results from both tests. J43001 seems to be a working part with inverting characteristics. J43000 does not show the same promise, it will need to be tested again to determine the reasoning behind our data. | + | * For the BBa_J43001, calculated error provides a possible explaination for why 100% hydrogen concentration fluoresced more than 75% hydrogen concentration. More testing required to determine the 100% hydrogen result. |
| - | * Further testing should be performed to determine if this promoter is actually an hydrogen sensing promoter, and therefore, an inverter. At the moment we do not have the ideal equipment for testing fluorescence. With more funding and better equipment, we might be able to get a better idea of the capabilities of our parts. | + | |
| | + | |
| | + | * Achievements: |
| | + | |
| | + | * The results of our test agree with possible success for part BBa_J43001. More testing must be performed to further prove our part as a hydrogen detector. |
| | + | * Our machine confirms the success of hybB, BBa_Q04121, and BBa_E0430 as a composite part |
| | + | |
| | + | |
| | |} | | |} |
| | | | |
The Team
Faculty Members:
- Dr. Filip To Agricultural and Biological Engineering
- Dr. Bob Reese Electrical and Computer Engineering
- Dr. Tod French Chemical Engineering
- Dr. Din-Pow Ma Biochemistry
Students:
- Teri Vaughn Undergraduate, Senior, Biomedical Engineering
- Courtney Harbin Undergraduate, Senior, Biochemistry
- Lauren Beatty Undergraduate, Junior, Biomedical Engineering
- Scott Tran Undergraduate, Junior, Biological Engineering
- Sam Pote Undergraduate, Freshman, Biological Engineering
- Paul Kimbrough Undergraduate, Junior, Biological Engineering
- Joseph Chen Undergraduate, Junior, Biological Engineering
- Robert Morris Grad student, Biological Engineering
- Meng-Hsuan Ho Grad student, Molecular Biology
- Brendan Flynn Grad student, Biological Engineering
|
Introduction
- International Genetically Engineered Machine (iGEM) is a student-led competition to build the most innovative "machine" by synthetic biology.
- Headquarters is located at the Massachusetts Institute of Technology.
- In 2006, 37 schools and over 400 students from around the world are participating in projects to construct biologically engineered systems.
- Task of each team is to apply engineering methodology to design and develop a new biological system ("machine") through the use of existing and/or newly formed microscopic biological parts (termed BioBricks).
- Type of the "machine" is chosen by each individual school participating, and the only criterion is that the "machine" be made entirely of the functional units of DNA called BioBricks.
- A registry of all BioBricks is kept in the MIT Registry of Standard Biological Parts, which is regularly updated to include new parts developed by teams.
- Parts for each iGEM team are obtained through the Registry for a fee.
- Jamboree for students to present their projects will take place at MIT in November.
|