|
|
| (10 intermediate revisions not shown) |
| Line 64: |
Line 64: |
| | | | |
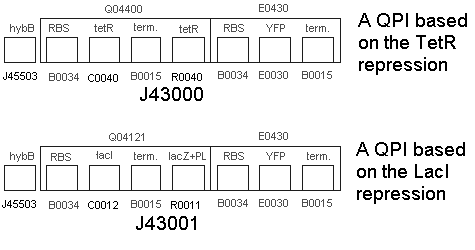
| | * We built the following two constructs:<br> | | * We built the following two constructs:<br> |
| - | :[[Image:untitled4.gif]] | + | :[[Image:untitled6.gif]] |
| | | | |
| | |} | | |} |
| Line 73: |
Line 73: |
| | <h3>Results</h3> | | <h3>Results</h3> |
| | | | |
| - | :[[Image:results.jpeg]] | + | :[[Image:MSUResultsErrorbars.JPG]] |
| - | |}
| + | |
| - | | + | |
| | | | |
| | * Part BBa_J43000 | | * Part BBa_J43000 |
| | | | |
| - | * No Hydrogen, No inducer = no fluorescence (except for a bit of leakage possibly). | + | * No Hydrogen, No tetR = fluorescence |
| - | * No Hydrogen, Inducer = Start Transcription at R0011 tetR, fluoresce. | + | * No Hydrogen, with tetR = Repressed Transcription at R0040 tetR, no fluorescence (except for some leakage). |
| - | * Hydrogen + Inducer = start transcription at hybB, produce tetR, competitive inhibition of operon at R0011 tetR. Less fluorescence transcribed. | + | * Hydrogen + tetR = start trqanscription at hybB, produce tetR, increased repression of operon at R0040 tetR. no fluorescence. |
| - | {| cellspacing="2px" cellpadding="0" border="0" style="padding: 0; width: 750px; color: #000000; background-color: #ffffff;"
| + | |
| - | | width=717px style="padding: 5px; background-color: #ffffff; border: 2px solid #993300;" |
| + | |
| | | | |
| - | <h3>Discussion</h3>
| |
| | | | |
| - | * Part BBa_J43000 | + | * Part BBa_J43001 |
| - |
| + | |
| | | | |
| - | * No Hydrogen, No inducer = no fluorescence (except for a bit of leakage possibly). | + | * No Hydrogen, No inducer = no fluorescence (except for a bit of leakage possibly). |
| - | * No Hydrogen, Inducer = Start Transcription at R0011 tetR, fluoresce. | + | * No Hydrogen, Inducer = Start Transcription at lacZ+PL, fluoresce. |
| - | * Hydrogen + Inducer = start transcription at hybB, produce tetR, competitive inhibition of operon at R0011 tetR. Less fluorescence transcribed. | + | * Hydrogen + Inducer = start transcription at hybB, produce lacI, competitive inhibition of operon at lacZ+PL. Less fluorescence transcribed. |
| | | | |
| - | * Complications – Bba_Q04400 was found out to be an inverter. This complicates our experiment and possibly explains the unusual results in fluorescence change for BBa_J43000. Further experimentation with this part might explain the inconsistemcies in the results.
| + | |} |
| | | | |
| | + | {| cellspacing="2px" cellpadding="0" border="0" style="padding: 0; width: 750px; color: #000000; background-color: #ffffff;" |
| | + | | width=717px style="padding: 5px; background-color: #ffffff; border: 2px solid #993300;" | |
| | | | |
| | + | <h3>Discussion</h3> |
| | | | |
| - | * Part BBa_J43001 | + | * Complications: |
| | + | |
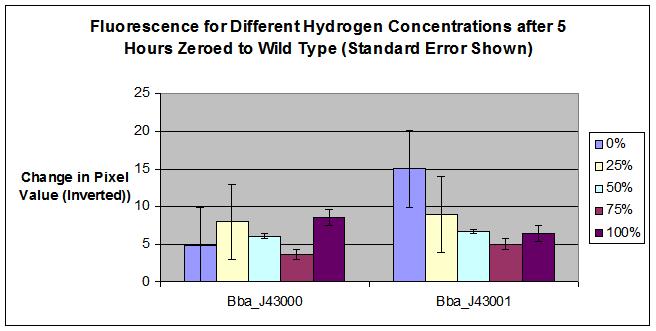
| | + | * BBa_J43000 was a failed part, we learned during completion of the construction and testing that there was a flaw in the design of the composite part. Although the hybB promoter does appear to detect hydrogen, the YFP is inhibited with the addition of tetR. Therefore, we cannot detect a change in fluorescence when hydrogen is added. |
| | + | * For the BBa_J43001, calculated error provides a possible explaination for why 100% hydrogen concentration fluoresced more than 75% hydrogen concentration. More testing required to determine the 100% hydrogen result. |
| | | | |
| | | | |
| - | * No Hydrogen, No inducer = no fluorescence (except for a bit of leakage possibly) | + | * Achievements: |
| - | * No Hydrogen, Inducer = Start Transcription at lacZ+PL, fluoresce
| + | |
| - | * Hydrogen + Inducer = start transcription at hybB, produce lacI, competitive inhibition of operon at lacZ+PL. Less fluorescence transcribed
| + | |
| | | | |
| - | * The results of our test agree with possible success for part BBa_J43001. More testing must be performed to further prove. Error bars should be added to possibly explain why 100% fluoresced more than 75%. Our machine confirms the success of hybB, BBa_Q04121, and BBa_E0430 | + | * The results of our test agree with possible success for part BBa_J43001. More testing must be performed to further prove our part as a hydrogen detector. |
| | + | * Our machine confirms the success of hybB, BBa_Q04121, and BBa_E0430 as a composite part |
| | | | |
| | | | |
The Team
Faculty Members:
- Dr. Filip To Agricultural and Biological Engineering
- Dr. Bob Reese Electrical and Computer Engineering
- Dr. Tod French Chemical Engineering
- Dr. Din-Pow Ma Biochemistry
Students:
- Teri Vaughn Undergraduate, Senior, Biomedical Engineering
- Courtney Harbin Undergraduate, Senior, Biochemistry
- Lauren Beatty Undergraduate, Junior, Biomedical Engineering
- Scott Tran Undergraduate, Junior, Biological Engineering
- Sam Pote Undergraduate, Freshman, Biological Engineering
- Paul Kimbrough Undergraduate, Junior, Biological Engineering
- Joseph Chen Undergraduate, Junior, Biological Engineering
- Robert Morris Grad student, Biological Engineering
- Meng-Hsuan Ho Grad student, Molecular Biology
- Brendan Flynn Grad student, Biological Engineering
|
Introduction
- International Genetically Engineered Machine (iGEM) is a student-led competition to build the most innovative "machine" by synthetic biology.
- Headquarters is located at the Massachusetts Institute of Technology.
- In 2006, 37 schools and over 400 students from around the world are participating in projects to construct biologically engineered systems.
- Task of each team is to apply engineering methodology to design and develop a new biological system ("machine") through the use of existing and/or newly formed microscopic biological parts (termed BioBricks).
- Type of the "machine" is chosen by each individual school participating, and the only criterion is that the "machine" be made entirely of the functional units of DNA called BioBricks.
- A registry of all BioBricks is kept in the MIT Registry of Standard Biological Parts, which is regularly updated to include new parts developed by teams.
- Parts for each iGEM team are obtained through the Registry for a fee.
- Jamboree for students to present their projects will take place at MIT in November.
|