ETH2006 iptg
From 2006.igem.org
(Difference between revisions)
(→Current implementation) |
|||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | →[[ETH Zurich 2006#System modeling|back to main page]] | ||
| + | |||
== Chemical sensing device == | == Chemical sensing device == | ||
| - | The chemical sensing device's | + | The chemical sensing device's PoPS output activity should be a monotonic function of the concentration of a certain substance in the cell's environment. |
=== Implementation alternatives === | === Implementation alternatives === | ||
| Line 21: | Line 23: | ||
=== Assembly procedure === | === Assembly procedure === | ||
| - | ''Where we got it and link to | + | ''Where we got it and link to their documentation'' |
Progress: | Progress: | ||
| Line 28: | Line 30: | ||
;2006/10/04: Transformed cells, plated them | ;2006/10/04: Transformed cells, plated them | ||
;2006/10/05: Found plates empty after 18h on the table, put into incubator at 37°C | ;2006/10/05: Found plates empty after 18h on the table, put into incubator at 37°C | ||
| - | ;2006/10/06: picked | + | ;2006/10/06: found lot of colonies, picked single ones and restreaked onto fresh plates |
| - | + | ||
| - | + | ||
=== Test procedure === | === Test procedure === | ||
| + | ;2006/10/07: streaked transformed bacteria on 4 different plates: | ||
| + | *plus IPTG, plus X-Gal | ||
| + | *plus IPTG, no X-Gal | ||
| + | *no IPTG, plus X-Gal | ||
| + | *no IPTG, no X-Gal | ||
| + | all plates contained the appropriate antibiotic (Chloramphenicol) | ||
=== Test results === | === Test results === | ||
| + | ;2006/10/08: results: | ||
| + | *plus IPTG, plus X-Gal: blue | ||
| + | *plus IPTG, no X-Gal: white | ||
| + | *no IPTG, plus X-Gal: light blue (system is leaky) | ||
| + | *no IPTG, no X-Gal: white | ||
| - | + | [[Image:chem_test.jpg|600px|center]] | |
| - | + | ||
| - | + | ||
Latest revision as of 12:41, 1 November 2006
Contents |
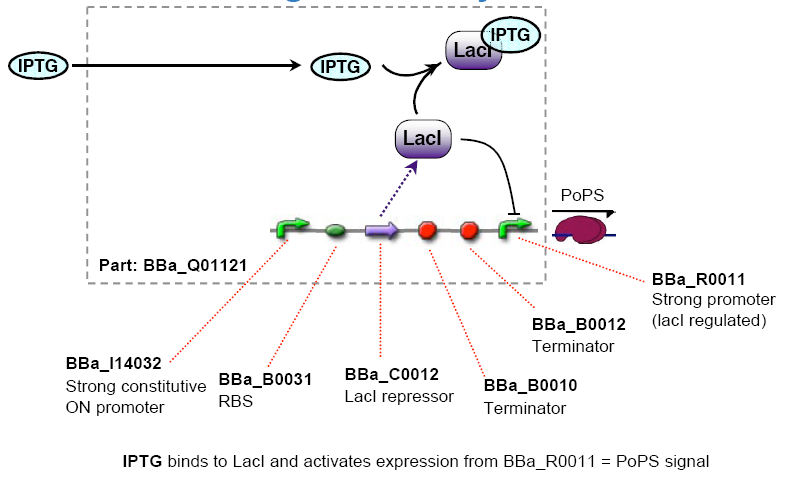
Chemical sensing device
The chemical sensing device's PoPS output activity should be a monotonic function of the concentration of a certain substance in the cell's environment.
Implementation alternatives
- Lactate lacI represses, IPTG induces ([http://partsregistry.org/Part:BBa_R0011 BBa_R0011] or [http://partsregistry.org/Part:BBa_R0010 BBa_R0010] )
- Tetracycline, TetR inhibitor, Tet inducer by inhibiting TetR (or aTc, it's analog) ([http://partsregistry.org/Part:BBa_R0040 BBa_R0040])
- combination thereof ([http://partsregistry.org/Part:BBa_I13614 BBa_I13614] / [http://partsregistry.org/Part:BBa_I13617 BBa_13617] / [http://partsregistry.org/Part:BBa_I13623 BBa_I13623] / [http://partsregistry.org/Part:BBa_I13624 BBa_I13624] / [http://partsregistry.org/Part:BBa_I13627 BBa_I13627] / [http://partsregistry.org/Part:BBa_I13637 BBa_I13637] / [http://partsregistry.org/Part:BBa_I13653 BBa_I13653])
- simple sugar Arabinose ([http://partsregistry.org/Part:BBa_R0080 BBa_R0080])
I see the main difficulty in the spatial separation as the cells are growing in the petri dishes. Since the inducers are water-soluble we would have to fix the chemicals onto the petri dish.
Current implementation
Since most parts were already existing in the registry, we decided to use the LacI system with IPTG as chemical to be sensed.
Modeling
Assembly procedure
Where we got it and link to their documentation
Progress:
- 2006/10/03
- Made LB-Agar plates with antibiotics
- 2006/10/04
- Transformed cells, plated them
- 2006/10/05
- Found plates empty after 18h on the table, put into incubator at 37°C
- 2006/10/06
- found lot of colonies, picked single ones and restreaked onto fresh plates
Test procedure
- 2006/10/07
- streaked transformed bacteria on 4 different plates:
- plus IPTG, plus X-Gal
- plus IPTG, no X-Gal
- no IPTG, plus X-Gal
- no IPTG, no X-Gal
all plates contained the appropriate antibiotic (Chloramphenicol)
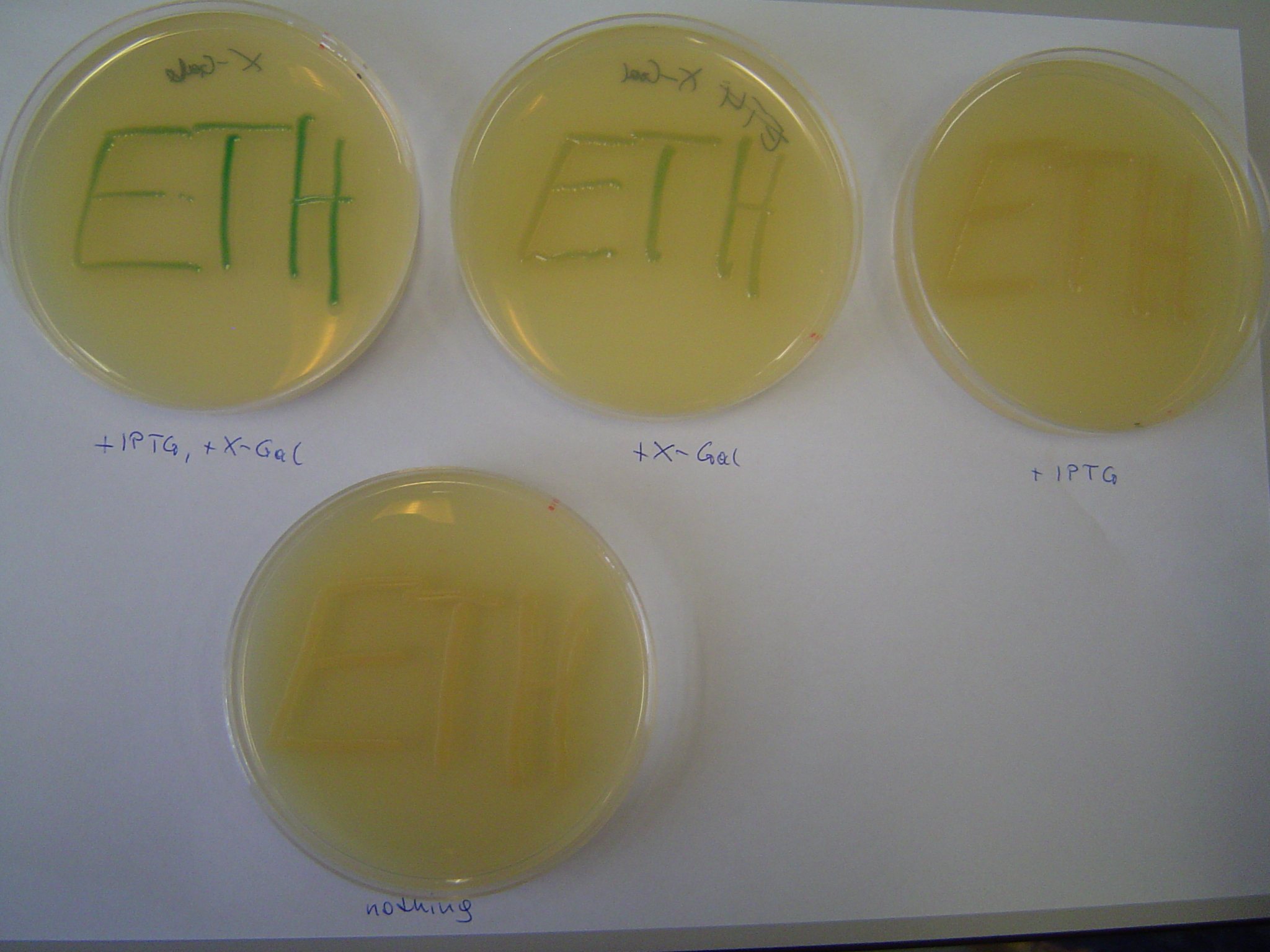
Test results
- 2006/10/08
- results:
- plus IPTG, plus X-Gal: blue
- plus IPTG, no X-Gal: white
- no IPTG, plus X-Gal: light blue (system is leaky)
- no IPTG, no X-Gal: white