Harvard 2006
From 2006.igem.org
(→Cell Surface Targeting) |
m (→The Harvard University 2006 iGEM Team: advisors -> advisers) |
||
| (58 intermediate revisions not shown) | |||
| Line 30: | Line 30: | ||
|} | |} | ||
| - | + | =The Harvard University 2006 iGEM Team= | |
| - | This year Harvard's team consisted of 10 undergraduate students, with backgrounds in molecular and cellular biology, biochemistry, and computer science. With the help of five faculty | + | This year Harvard's team consisted of 10 undergraduate students, with backgrounds in molecular and cellular biology, biochemistry, and computer science. With the help of five faculty advisers and three graduate-level teaching advisers, they devised and executed three separate projects. |
| - | 1. Construction of novel DNA nanostructures for the purpose of stealth drug delivery | + | 1. [[#DNA_Nanobox:_A_Nanoscale_Device_for_Stealth_Drug_Delivery | Construction of novel DNA nanostructures for the purpose of stealth drug delivery.]] |
| - | 2. Exploration of cell | + | 2. [[#Cell_Surface_Targeting | Exploration of cell surface targeting by using interchangable and linkable aptamers (adaptamers), and an Lpp-OmpA fusion protein for surface-displayed streptavidin.]] |
| - | 3. Reconstitution of a circadian oscillator from cyanobacteria into E. coli | + | 3. [[#A_Circadian_Oscillator_for_E.coli | Reconstitution of a circadian oscillator from cyanobacteria into E. coli.]] |
| - | + | =DNA Nanobox: A Nanoscale Device for Stealth Drug Delivery= | |
| - | + | ||
| - | + | ||
'''Project Overview''' | '''Project Overview''' | ||
| - | *Traditional drug delivery is | + | *Traditional drug delivery is fairly low-tech. Tablets containing a mixture of active small molecules and binders are ingested, and the small molecules circulate non-specifically throughout the body. Even when a specific cell or tissue type is targeted (e.g. cancerous cells or bacterial infections), many other tissues are exposed to the drugs, often with serious side effects. We envision a future in which medicines will be delivered in a highly specific manner to targeted tissues and cell types, and set out to make initial steps toward realizing this vision. |
*Our goal for the summer was to design and implement nanoscale molecular containers, which could be dynamically opened and closed by an external stimulus. This is the first step to making more sophisticated drug-delivery vehicles. | *Our goal for the summer was to design and implement nanoscale molecular containers, which could be dynamically opened and closed by an external stimulus. This is the first step to making more sophisticated drug-delivery vehicles. | ||
*The containers were implemented as DNA nanostructures, which afford a significant degree of positional control and chemical versatility. | *The containers were implemented as DNA nanostructures, which afford a significant degree of positional control and chemical versatility. | ||
| Line 51: | Line 49: | ||
'''Results''' | '''Results''' | ||
| - | *The following images show our most successful container design, both as a 3D cartoon and in actual negative-stain EM images. The length, width, and depth of this container design is approximately 30x30x30 nanometers, so it is bigger than a typical ribosome. | + | *The following images show our most successful container design, both as a 3D cartoon and in actual negative-stain EM images. The outer length, width, and depth of this container design is approximately 30x30x30 nanometers, so it is bigger than a typical ribosome. The inner diameter of the container is approximately 12 nanometers. |
<gallery> | <gallery> | ||
Image:Harvard2006-nanobox-3Dview1.jpg|3D Cross Section | Image:Harvard2006-nanobox-3Dview1.jpg|3D Cross Section | ||
| Line 71: | Line 69: | ||
*[http://openwetware.org/wiki/IGEM:Harvard/2006/DNA_nanostructures openwetware page] | *[http://openwetware.org/wiki/IGEM:Harvard/2006/DNA_nanostructures openwetware page] | ||
| - | + | =Cell Surface Targeting= | |
| - | + | ==Adaptamers== | |
'''Project Overview''' | '''Project Overview''' | ||
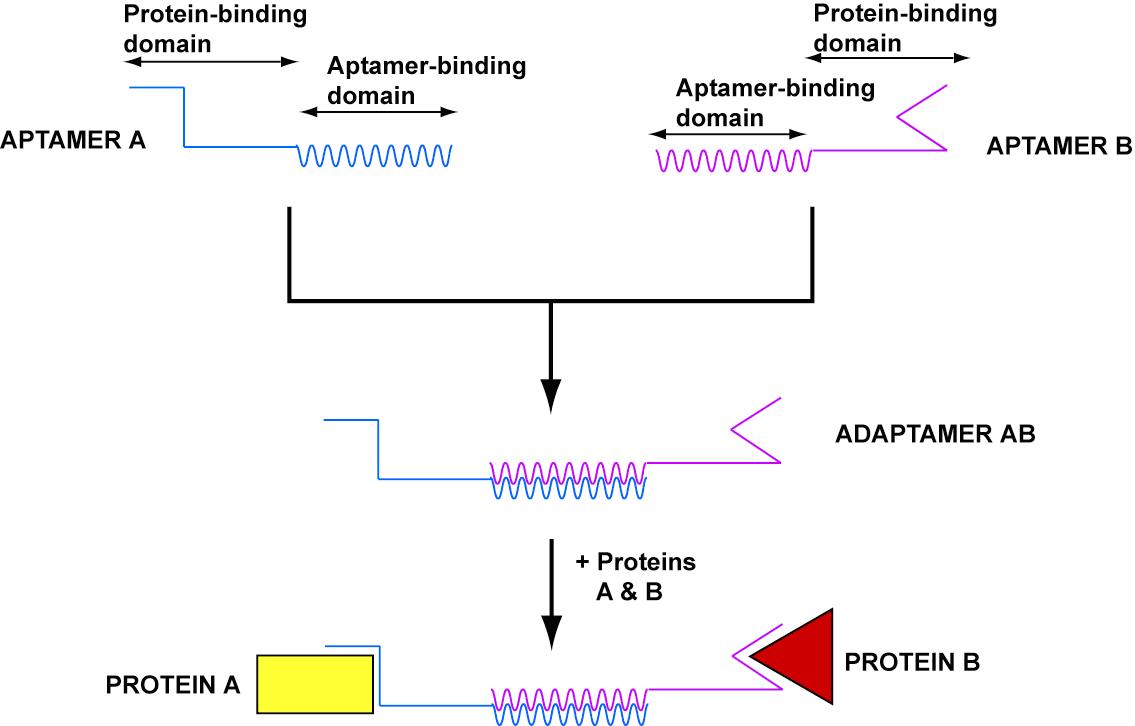
*In order to target nanostructures to cells, we developed adaptamers, universal nucleic acid adaptars which can link two substrates. | *In order to target nanostructures to cells, we developed adaptamers, universal nucleic acid adaptars which can link two substrates. | ||
| Line 78: | Line 76: | ||
*Adaptamers generally depend on aptamers, short sequences of nucleic acid that bind with high specificity and affinity to particular substrates. | *Adaptamers generally depend on aptamers, short sequences of nucleic acid that bind with high specificity and affinity to particular substrates. | ||
*Tahiri-Alaoui et al. created the first aptamer in 2002, consisting of two aptamer sequences linked together by a bulky basepairing region ~100 nucleotides long. | *Tahiri-Alaoui et al. created the first aptamer in 2002, consisting of two aptamer sequences linked together by a bulky basepairing region ~100 nucleotides long. | ||
| - | *Our goal was to create an adaptamer that could link together streptavidin and thrombin. Delivery of thrombin to a streptavidin-coated magnetic bead would show the potential for delivery of a macromolecule to a cell surface. | + | *Our goal was to create an adaptamer that could link together streptavidin and thrombin. Delivery of thrombin to a streptavidin-coated magnetic bead would show the potential for delivery of a macromolecule to a cell surface (see gallery). |
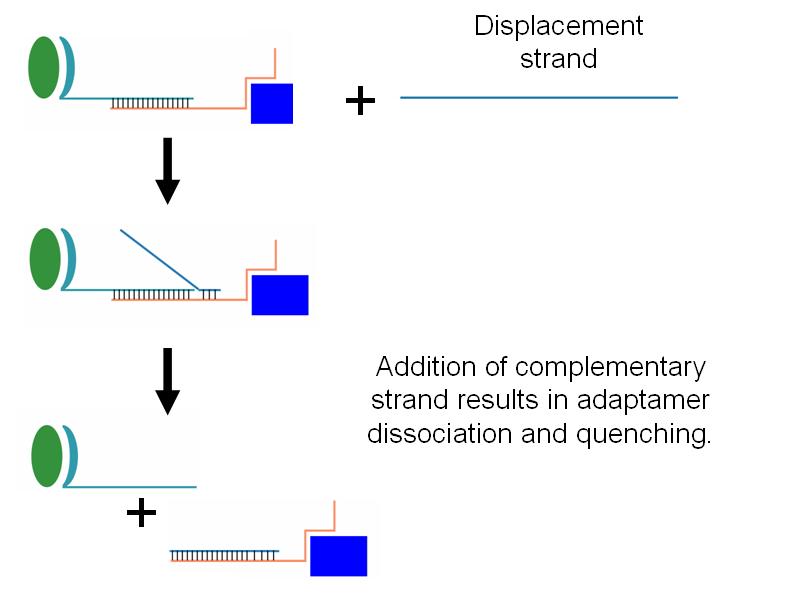
| - | *Additionally, we wished to be able to be able to quench adaptamer function through the addition of an adapatamer-disabling oligonucleotide. | + | *Additionally, we wished to be able to be able to quench adaptamer function through the addition of an adapatamer-disabling oligonucleotide (see gallery). |
| + | |||
| + | <gallery> | ||
| + | Image:Harvard2006-adaptamer-creation.jpg|Adaptamer design | ||
| + | Image:Harvard2006-stranddisplacement.jpg|Strand displacement | ||
| + | </gallery> | ||
'''Results''' | '''Results''' | ||
*Our design consisted of a streptavidin aptamer and thrombin aptamer linked together by a short, variable-length double-stranded region of linker DNA. | *Our design consisted of a streptavidin aptamer and thrombin aptamer linked together by a short, variable-length double-stranded region of linker DNA. | ||
*In order to prevent interaction of linker DNA with aptamers, we wrote a program that can evolve a sequence for the linking region given two aptamer sequences. This program can be used for any aptamer sequences and will be utilized for future adaptamer designs. | *In order to prevent interaction of linker DNA with aptamers, we wrote a program that can evolve a sequence for the linking region given two aptamer sequences. This program can be used for any aptamer sequences and will be utilized for future adaptamer designs. | ||
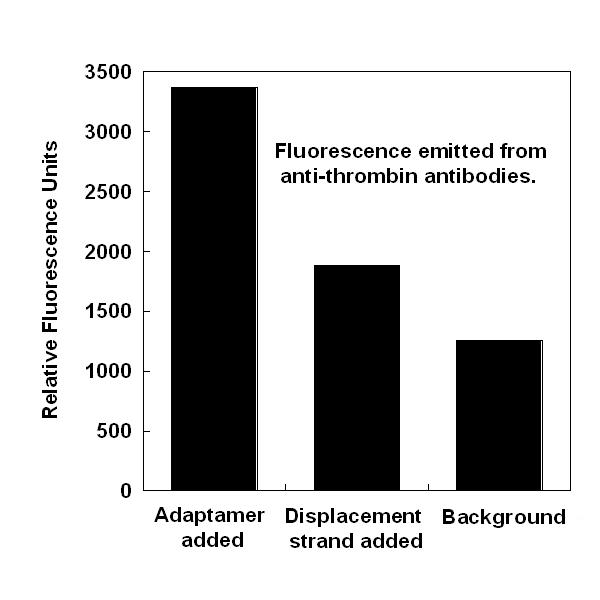
| - | *We successfully showed that four separate adaptamer designs are capable of targeting thrombin to streptavidin-coated magnetic beads by probing targeted beads with thrombin antibodies. | + | *We successfully showed that four separate adaptamer designs are capable of targeting thrombin to streptavidin-coated magnetic beads by probing targeted beads with thrombin antibodies (see gallery). |
| - | *We successfully showed that addition of a displacement strand which breaks apart adaptamers was capable of quenching adaptamer function. | + | *We successfully showed that addition of a low concentration of displacement strand which breaks apart adaptamers was capable of quenching adaptamer function (see gallery). |
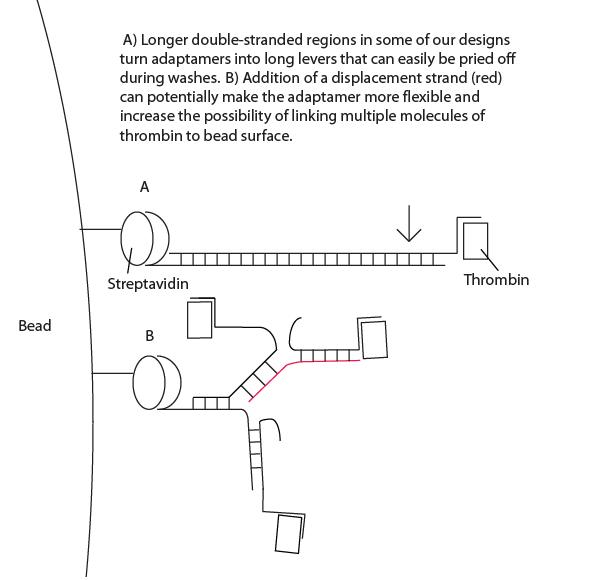
| - | *Interestingly, high concentrations of displacement strand led to greater maintenance of thrombin targeting. We have proposed a lever-hypothesis for this observation. | + | *Interestingly, high concentrations of displacement strand led to greater maintenance of thrombin targeting. See Project Notebook for details. We have proposed a lever-hypothesis for this observation (see gallery). |
| + | |||
| + | <gallery> | ||
| + | Image:Harvard2006-BeadAssay.jpg|Adaptamers successfully targeted thrombin to streptavidin-coated beads. | ||
| + | Image:Harvard2006-StrandDisplacementresult.jpg|Strand displacement successfully quenched adaptamer function. | ||
| + | Image:Harvard2006-Lever-hyp.jpg|Lever-hypothesis | ||
| + | </gallery> | ||
'''Future Plans''' | '''Future Plans''' | ||
| - | *We will demonstrate the modular design of adaptamers by using an adaptamer to target thrombin to | + | *We will demonstrate the modular design of adaptamers by using an adaptamer to target thrombin to Jurkat cells. |
*We will attempt to prove the lever-hypothesis. | *We will attempt to prove the lever-hypothesis. | ||
'''Electronic Notebook''' | '''Electronic Notebook''' | ||
| - | *[http://openwetware.org/wiki/IGEM:Harvard/2006/Adaptamers openwetware page] | + | *[http://openwetware.org/wiki/IGEM:Harvard/2006/Adaptamers/Overview openwetware page] |
| + | ==Cell surface streptavidin== | ||
| + | '''Project Overview''' | ||
| + | *While the adaptamer project uses nucleic acids to target any two substrates to each other, the cell surface streptavidin project seeks to target any substrate(s) to the cell surface. | ||
| + | *The method of targeting would be implemented through the '''expression of streptavidin protein on the cell surface'''. Streptavidin is a protein which binds strongly to the biotin molecule. We would be utilizing this strong streptavidin-biotin binding to target <u>any</u> biotinylated nucleic acid or peptide to the outside of a cell expressing streptavidin on the surface. | ||
| + | *Some potential applications include bringing a biotinylated protein to the cell surface for facilitated interaction with some other surface element, linking a cell with another protein/cell via a biotinylated aptamer, or delivering a DNA nanobox to the cell surface through a biotinylated oligonucleotide. | ||
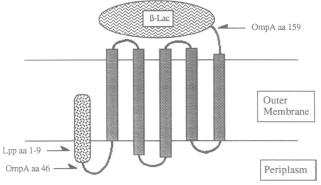
| - | + | *Streptavidin would be expressed on the cell surface through the '''Lpp-OmpA surface display vehicle''' (Earhart, 2000). The complete vehicle consists of three fused protein domains: signal peptide of lipoprotein (Lpp), transmembrane domains of outer membrane protein A (OmpA), and the surface protein. | |
| - | * | + | |
| - | + | ||
| - | ''' | + | *Assembly of the fusion protein would be carried out through a '''modified BioBricks assembly for protein domains''' (Phillips and Silver, 2006). Standard BioBricks parts separated from the flanking XbaI and SpeI sites by a single spacer nucleotide, in order to prevent Dam methylation. If you leave out these spacer nucleotides, the mixed site formed between assembled parts is six base pairs long, and so reading frame can be maintained between assembled protein domains. |
| + | |||
| + | <gallery> | ||
| + | Image:Lpp ompa.JPG|Lpp-OmpA surface display vehicle (Francisco et al., 1993) | ||
| + | Image:Protein domain biobrick.JPG|Modified BioBricks assembly for protein domains | ||
| + | </gallery> | ||
'''Results''' | '''Results''' | ||
| + | *We completed assembly of constructs using the following BioBrick parts. | ||
| + | **<bbpart>BBa_J04500</bbpart>. a composite part of a lac promoter (R0010) and a strong ribosome binding site (B0034) | ||
| + | **<bbpart>BBa_J36835</bbpart>. Lpp, the lipoprotein signal peptide. | ||
| + | **<bbpart>BBa_J36837</bbpart> or <bbpart>BBa_J36838</bbpart>. OmpA, one (O1) or five (O5) transmembrane domains, respectively. Both have been shown to work (Earhart, 2000). | ||
| + | **<bbpart>BBa_J36841</bbpart> or <bbpart>BBa_J36843</bbpart>. Streptavidin, either wild-type "SW" (Howarth, 2006), or single-chain dimeric "SD" (Aslan, 2005). | ||
| + | ***Note: Streptavidin exists naturally as a soluble tetramer, but restriction to the surface might allow only monomeric/dimeric forms. What is interesting is that there has been research done to engineer such forms, in order to render biotin binding less strong but more easily reversible (Wu, 2005). | ||
| + | *Western blots were performed with transformed and induced cells, probing with anti-his6 antibody (each streptavidin had a His6 tag) and with anti-streptavidin antibody. Distinct bands were observed at the expected sizes in anti-his6 probing, and bands were observed at the same places with the anti-streptavidin probing. These results suggest the following. | ||
| + | **The promoter and ribosome binding site are functioning correctly for expression of these fusion protein construct. | ||
| + | **The BioBricks assembly of the fusion protein was successful in that reading frame was maintained, since the coding sequence for the His6 tag is found at the end of the construct. | ||
| + | **The streptavidin part of the fusion protein is still folding in such a way that anti-streptavidin antibody can recognize it. | ||
| + | |||
| + | <gallery> | ||
| + | Image:Cell surface streptavidin construct.JPG|Diagram of cell surface streptavidin construct | ||
| + | Image:Cell surface streptavidin westerns.JPG|Western blots of cell surface streptavidin constructs | ||
| + | </gallery> | ||
'''Future Plans''' | '''Future Plans''' | ||
| + | *Now that we know that the construct is being expressed, we need to determine whether or not the construct is being localized to the outer membrane. We can separate the cell lysate by centrifugation, isolate the outer membrane proteins, and then perform Western blots, probing with anti-his6 and anti-streptavidin antibody. | ||
| + | *If the construct is indeed being localized to the outer membrane, we then need to determine if the streptavidin is being displayed functionally on the cell surface. We can probe whole cells with anti-streptavidin antibodies or even fluorescently tagged streptavidin aptamers for in-cell westerns, or we can visualize surface binding of biotinylated, fluorescently tagged oligonucleotides under a microscope. | ||
| + | *If streptavidin is being expressed on the cell surface, we can switch in other engineered streptavidin clones and compare biotin binding. We can also try adding a length of amino acids between OmpA and streptavidin, which might give spatial flexibility to allow formation of tetramers on the cell surface or to allow the streptavidin to extend outside of any extracellular complexes. | ||
'''Electronic Notebook''' | '''Electronic Notebook''' | ||
*[http://openwetware.org/wiki/IGEM:Harvard/2006/Fusion_proteins openwetware page] | *[http://openwetware.org/wiki/IGEM:Harvard/2006/Fusion_proteins openwetware page] | ||
| - | + | '''References''' | |
| + | *Francisco JA, Earhart CF, and Georgiou G. Transport and anchoring of beta-lactamase to the external surface of Escherichia coli. Proc Natl Acad Sci U S A 1992 Apr 1; 89(7) 2713-7. | ||
| + | *Earhart CF. Use of an Lpp-OmpA fusion vehicle for bacterial surface display. Methods Enzymol 2000; 326 506-16. | ||
| + | *Aslan FM, Yu Y, Mohr SC, and Cantor CR. Engineered single-chain dimeric streptavidins with an unexpected strong preference for biotin-4-fluorescein. Proc Natl Acad Sci U S A 2005 Jun 14; 102(24) 8507-12. | ||
| + | *Wu SC and Wong SL. Engineering soluble monomeric streptavidin with reversible biotin binding capability. J Biol Chem 2005 Jun 17; 280(24) 23225-31. | ||
| + | *Howarth M, Chinnapen DJ, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, El-Husseini A, and Ting AY. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods 2006 Apr; 3(4) 267-73. | ||
| + | *Phillips I, Silver P, A New Biobrick Assembly Strategy Designed for Facile Protein Engineering, MIT Synthetic Biology Working Group Reports, published online 20 April 2006 at [https://dspace.mit.edu/handle/1721.1/32535 MIT dspace]. | ||
| + | =A Circadian Oscillator for E.coli= | ||
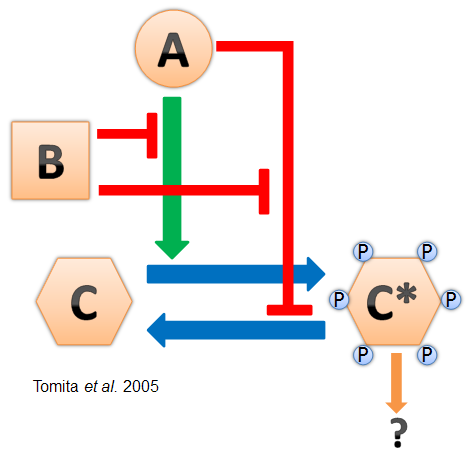
| + | [[Image:KaiProteinInteraction.png|thumb|left|'''One proposed model of the Kai protein interactions.'''<br>'''KaiA''' promotes the autosphorylation and inhibits the autodephosphorylation of '''KaiC.'''<br> '''KaiB''' inhibits the action of '''KaiA.''' ''Tomita ''et al'', 2005'']] | ||
'''Project Overview''' | '''Project Overview''' | ||
| + | *Oscillators are used in a variety of electronic circuits and would be essential in complex, time-dependant biocircuitry. The synthetic "repressilator" system engineered by Elowitz et al. (2000), though a good proof of concept, lacks the stability needed for long-term usage. | ||
| + | *Cyanobacteria have a naturally-evolved oscillator driven by the interactions between three proteins named KaiA, KaiB, and KaiC (see diagram at left). This "Kai clock" does not require transcriptional regulation and has been found to oscillate in vitro (Nakajima et al. 2005). It is also robust and can be modified for longer and shorter periods by point mutations in the Kai genes (Kondo et al. 1994). | ||
| + | |||
| + | *Based on these discoveries, our goal for the summer was to reconstitute the Kai oscillation cycle in ''E. coli''. This would be the first step in creating a new oscillator part for the registry, as well as providing an important breakthrough for circadian biology research. | ||
| + | *The transcription factors that link the Kai clock to gene regulation in cyanobacteria are not well understood. Therefore we tested our clock in ''E. coli'' by measuring the amounts of phosphorylated and unphosphorylated KaiC via Western blot. | ||
'''Results''' | '''Results''' | ||
| + | *We have succesfully created BioBricks of the Kai genes and several functional constructs. These include: | ||
| + | **Standalone KaiA , KaiB , and KaiC as Biobricks.(<bbpart>BBa_J36801</bbpart>, <bbpart>BBa_J36802</bbpart>, <bbpart>BBa_J36804</bbpart>) | ||
| + | **Kai proteins with <bbpart>BBa_J04500</bbpart>, which consists of a Lac promoter + RBS. (<bbpart>BBa_J36831</bbpart>, <bbpart>BBa_J36832</bbpart>, <bbpart>BBa_J36834</bbpart>) | ||
| + | **KaiA/J04500 + KaiC/J04500 in order to test KaiA's effect on KaiC (which, according to the model, should be promotion of KaiC's phosphorylation). (<bbpart>BBa_J36335</bbpart>) | ||
| + | **KaiB/J04500 + KaiC/J04500 in order to test KaiB's effect on KaiC (which, according to the model, should be nothing). (<bbpart>BBa_J36336</bbpart>) | ||
| + | <gallery> | ||
| + | Image:KaiAKaiBKaiC.PNG|'''KaiA, KaiB, KaiC'''<Br> Silent point mutations allow Biobrick incorporation | ||
| + | Image:KaiProteinsWithJ04500.PNG|'''Kai genes with BBa_J04500''' <Br> Allow inducible expression in E. coli | ||
| + | Image:KaiAKaiC.PNG|'''KaiA/J04500 + KaiC/J04500''' <br> Used to test KaiA - KaiC post-transcriptional interaction in E. coli | ||
| + | Image:KaiBKaiC.PNG|'''KaiB/J04500 + KaiC/J04500''' <br> Used to test KaiB - KaiC post-transcriptional interaction in E. coli | ||
| + | </gallery> | ||
| + | |||
| + | *Based on the Tomita ''et al.'' 2005 model, we were able to develop hypotheses to predict the relative levels of phosphorylated/dephosphorylated KaiC in the presence of either KaiA or KaiB alone. Using the constructs we created, we tested and confirm these hypotheses. Anti-KaiC antibodies revealed a change in the relative concentration of dephosphorylated and phosphorylated KaiC from our different constructs. | ||
| + | *'''Hypotheses:''' | ||
| + | **KaiC alone will produce phosphorylated and dephosphorylated KaiC. This is based on the theory that KaiC autophosphorylates and autodephosphorylates. | ||
| + | **KaiA and KaiC will produce more phosphorylated than dephosphorylated KaiC, since KaiA promotes KaiC phosphorylation. | ||
| + | **KaiB will have no affect on KaiC's phosphorylation, since it does not interact with it directly according to the model. Therefore, the relative amounts of phosphorylated and dephosphorylated KaiC should be the same as when KaiC is expressed by itself. | ||
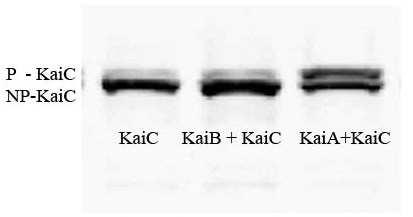
| + | [[Image:KaiCWesternBlot.jpg|thumb|right|'''A Western blot using anti-KaiC antibodies.''' The relative amount of phosphorylated KaiC is higher in the presence of KaiA than alone or in the presence of KaiB. This supports our predictions based on the described model.]] | ||
| + | *'''Western Blot results''' | ||
| + | **We performed a Western blot on our KaiC/J04500, KaiB/J04500 + KaiC/J04500, and KaiA/J04500 + KaiC/J04500 constructs using KaiC antibodies. Each sample was taken at the same OD. The results confirmed all of our hypotheses. | ||
| + | **In Lane 1 (KaiC/J04500), we can see a strong band of dephosphorylated KaiC and a weaker band of phosphorylated KaiC. This confirms that KaiC autophosphorylates and autodephosphorylates. | ||
| + | **In Lane 2(KaiB/J04500 + KaiC/J04500), we can again see a strong band of dephosphorylated KaiC and a weaker band of phosphorylated KaiC, in fact very similar, if not identical, to the results in Lane 1. This confirms that KaiB has no direct affect on KaiC's autophosphorylation, as predicted by the model. | ||
| + | **In Lane 3(KaiA/J04500 + KaiC/J04500), we see a strong band of dephosphorylated KaiC, but also a strong band of phosphorylated KaiC. Since the amount of phosphorylated KaiC is greater than that shown in Lanes 1 and 2, we have evidence that KaiA is interacting with KaiC to produce a greater amount of phosphorylated KaiC. | ||
| + | ** in summary, we were able to express the three Kai proteins in ''E. coli'', and show the predicted interaction between KaiA and KaiC and possible interaction between KaiB and KaiC. | ||
'''Future Plans''' | '''Future Plans''' | ||
| - | + | *Short term plans include replicating our previous Western blot results as well as further characterizing post-transcriptional interactions between KaiA-KaiC and KaiB-KaiC pairs. | |
| + | *In cyanobacteria, the Kai clock is reset by light/dark transitions. This serves to synchronize a population's circadian rhythms with the day/night cycle. This synchronization is necissary for experimenters to detect KaiC oscillation; a mixed-phase population will show no bulk, time-dependant change in KaiC phosphorylation state. Unfortunately, the molecular mechanism behind light/dark synchronization is not yet well understood, so we cannot transfer it to ''E.coli''. Instead, we have designed a set of experiments using inducible promoters for the Kai genes to synchronize the Kai clocks within each cell and between cells in a culture . Our goal is to coerce a culture of ''E.coli'' cells into synchronized oscillation, which we can then measure by Western blot. | ||
| + | *As more is discovered about the intermediate factors between the Kai clock and gene regulation in cyanobacteria, we would like to leverage these findings to make the Kai clock drive other components in a biological circuit. | ||
'''Electronic Notebook''' | '''Electronic Notebook''' | ||
*[http://openwetware.org/wiki/IGEM:Harvard/2006/Cyanobacteria openwetware page] | *[http://openwetware.org/wiki/IGEM:Harvard/2006/Cyanobacteria openwetware page] | ||
| - | |||
| - | |||
==Complete Electronic Notebooks== | ==Complete Electronic Notebooks== | ||
Detailed records of our summer activities, including results, can be found on openwetware, which the team used to host its [http://openwetware.org/wiki/IGEM:Harvard/2006 electronic notebook]. Links to specific project pages can be found in the above descriptions. | Detailed records of our summer activities, including results, can be found on openwetware, which the team used to host its [http://openwetware.org/wiki/IGEM:Harvard/2006 electronic notebook]. Links to specific project pages can be found in the above descriptions. | ||
Latest revision as of 20:57, 8 December 2006
| Harvard University 2006 iGEM Team | ||
Students
|
Teaching Fellows
Faculty Advisors
| |
The Harvard University 2006 iGEM Team
This year Harvard's team consisted of 10 undergraduate students, with backgrounds in molecular and cellular biology, biochemistry, and computer science. With the help of five faculty advisers and three graduate-level teaching advisers, they devised and executed three separate projects.
1. Construction of novel DNA nanostructures for the purpose of stealth drug delivery.
3. Reconstitution of a circadian oscillator from cyanobacteria into E. coli.
DNA Nanobox: A Nanoscale Device for Stealth Drug Delivery
Project Overview
- Traditional drug delivery is fairly low-tech. Tablets containing a mixture of active small molecules and binders are ingested, and the small molecules circulate non-specifically throughout the body. Even when a specific cell or tissue type is targeted (e.g. cancerous cells or bacterial infections), many other tissues are exposed to the drugs, often with serious side effects. We envision a future in which medicines will be delivered in a highly specific manner to targeted tissues and cell types, and set out to make initial steps toward realizing this vision.
- Our goal for the summer was to design and implement nanoscale molecular containers, which could be dynamically opened and closed by an external stimulus. This is the first step to making more sophisticated drug-delivery vehicles.
- The containers were implemented as DNA nanostructures, which afford a significant degree of positional control and chemical versatility.
- To demonstrate that our designs successfully self-assembled, we used negative-stain electron microscopy to visualize the containers.
- As an initial proof-of-concept, we planned to demonstrate that our DNA containers could be used to "protect" biotinylated oligonucleotides from binding by streptavidin-coated magnetic beads.
- We were able to successfully show that nanoboxes with internal biotinylated oligonucleotides did not bind to streptavidin-coated magnetic beads, while nanoboxes that were externally decorated with biotinylated oligonucleotides were bound by the same beads.
Results
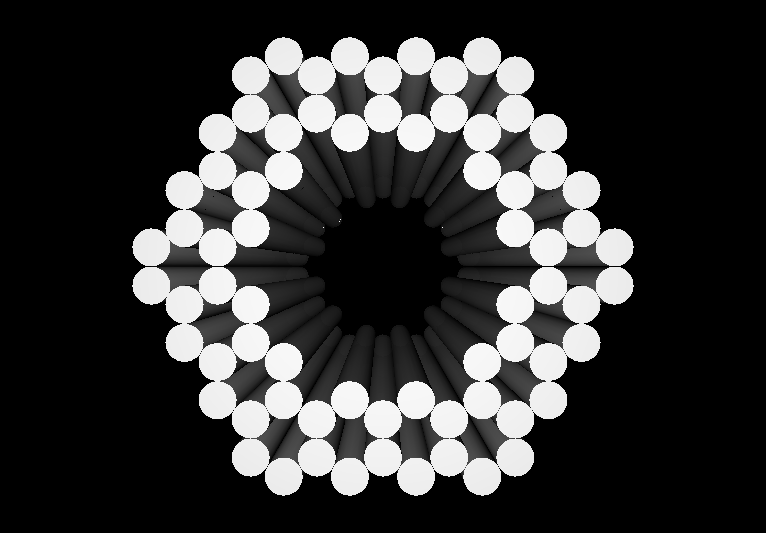
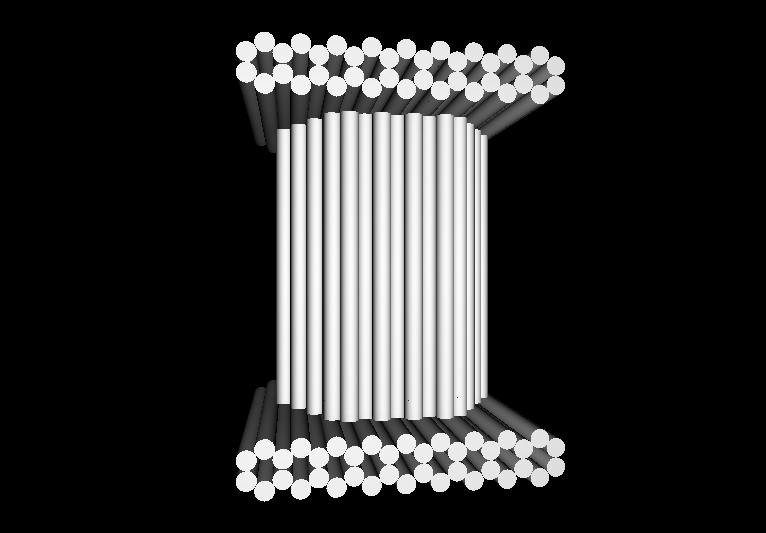
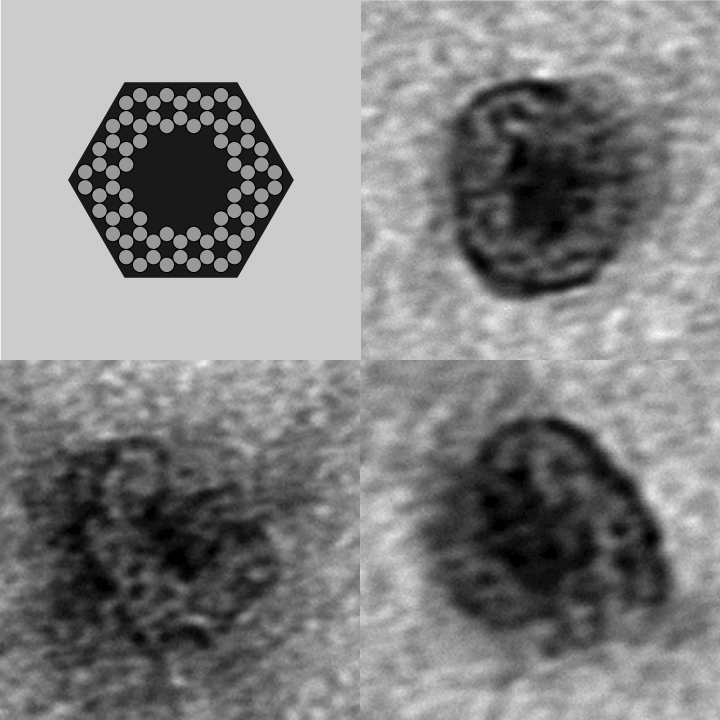
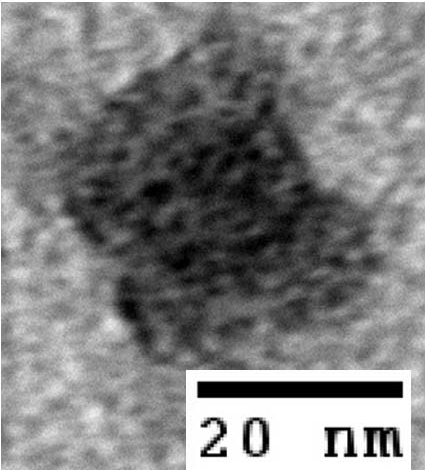
- The following images show our most successful container design, both as a 3D cartoon and in actual negative-stain EM images. The outer length, width, and depth of this container design is approximately 30x30x30 nanometers, so it is bigger than a typical ribosome. The inner diameter of the container is approximately 12 nanometers.
- More details about this design can be found [http://openwetware.org/wiki/IGEM:Harvard/2006/Container_Design_5 here].
- The basic design of this container is a hexagonal barrel, closed off by two flat lids.
- The lids are designed to reversibly attach to the barrel using complementary oligonucleotides using a "toehold" strategy in which the strands include unpaired overhangs that can be displaced with the addition of oligos that match for the full length of the sequence.
Future Plans
- We have demonstrated the first steps toward building a sophisticated, versatile drug delivery vehicle out of DNA.
- Short term plans include assembly of closed containers (which include lids), and further characterization of assembled structures (both imaging and functional assays). We have not determined the maximum size for a molecule that can still diffuse between DNA helices that comprise our container, but for a closed container we expect it to be around 1 nanometer in diameter.
- In the long term, we envision designing containers that incorporate cell-surface targeting mechanisms that would allow delivery of a payload to a specific cell type. Advances also need to be made if the container is going to deliver small molecules - perhaps by encapsulating the container with a liposome (or vice versa).
Electronic Notebook
- [http://openwetware.org/wiki/IGEM:Harvard/2006/DNA_nanostructures openwetware page]
Cell Surface Targeting
Adaptamers
Project Overview
- In order to target nanostructures to cells, we developed adaptamers, universal nucleic acid adaptars which can link two substrates.
- Such an interface could also be used to link together entire cells for the study of cell-cell interactions and the linkage of two interacting proteins, in effect creating a nucleic acid enzyme.
- Adaptamers generally depend on aptamers, short sequences of nucleic acid that bind with high specificity and affinity to particular substrates.
- Tahiri-Alaoui et al. created the first aptamer in 2002, consisting of two aptamer sequences linked together by a bulky basepairing region ~100 nucleotides long.
- Our goal was to create an adaptamer that could link together streptavidin and thrombin. Delivery of thrombin to a streptavidin-coated magnetic bead would show the potential for delivery of a macromolecule to a cell surface (see gallery).
- Additionally, we wished to be able to be able to quench adaptamer function through the addition of an adapatamer-disabling oligonucleotide (see gallery).
Results
- Our design consisted of a streptavidin aptamer and thrombin aptamer linked together by a short, variable-length double-stranded region of linker DNA.
- In order to prevent interaction of linker DNA with aptamers, we wrote a program that can evolve a sequence for the linking region given two aptamer sequences. This program can be used for any aptamer sequences and will be utilized for future adaptamer designs.
- We successfully showed that four separate adaptamer designs are capable of targeting thrombin to streptavidin-coated magnetic beads by probing targeted beads with thrombin antibodies (see gallery).
- We successfully showed that addition of a low concentration of displacement strand which breaks apart adaptamers was capable of quenching adaptamer function (see gallery).
- Interestingly, high concentrations of displacement strand led to greater maintenance of thrombin targeting. See Project Notebook for details. We have proposed a lever-hypothesis for this observation (see gallery).
Future Plans
- We will demonstrate the modular design of adaptamers by using an adaptamer to target thrombin to Jurkat cells.
- We will attempt to prove the lever-hypothesis.
Electronic Notebook
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Adaptamers/Overview openwetware page]
Cell surface streptavidin
Project Overview
- While the adaptamer project uses nucleic acids to target any two substrates to each other, the cell surface streptavidin project seeks to target any substrate(s) to the cell surface.
- The method of targeting would be implemented through the expression of streptavidin protein on the cell surface. Streptavidin is a protein which binds strongly to the biotin molecule. We would be utilizing this strong streptavidin-biotin binding to target any biotinylated nucleic acid or peptide to the outside of a cell expressing streptavidin on the surface.
- Some potential applications include bringing a biotinylated protein to the cell surface for facilitated interaction with some other surface element, linking a cell with another protein/cell via a biotinylated aptamer, or delivering a DNA nanobox to the cell surface through a biotinylated oligonucleotide.
- Streptavidin would be expressed on the cell surface through the Lpp-OmpA surface display vehicle (Earhart, 2000). The complete vehicle consists of three fused protein domains: signal peptide of lipoprotein (Lpp), transmembrane domains of outer membrane protein A (OmpA), and the surface protein.
- Assembly of the fusion protein would be carried out through a modified BioBricks assembly for protein domains (Phillips and Silver, 2006). Standard BioBricks parts separated from the flanking XbaI and SpeI sites by a single spacer nucleotide, in order to prevent Dam methylation. If you leave out these spacer nucleotides, the mixed site formed between assembled parts is six base pairs long, and so reading frame can be maintained between assembled protein domains.
Results
- We completed assembly of constructs using the following BioBrick parts.
- BBa_J04500. a composite part of a lac promoter (R0010) and a strong ribosome binding site (B0034)
- BBa_J36835. Lpp, the lipoprotein signal peptide.
- BBa_J36837 or BBa_J36838. OmpA, one (O1) or five (O5) transmembrane domains, respectively. Both have been shown to work (Earhart, 2000).
- BBa_J36841 or BBa_J36843. Streptavidin, either wild-type "SW" (Howarth, 2006), or single-chain dimeric "SD" (Aslan, 2005).
- Note: Streptavidin exists naturally as a soluble tetramer, but restriction to the surface might allow only monomeric/dimeric forms. What is interesting is that there has been research done to engineer such forms, in order to render biotin binding less strong but more easily reversible (Wu, 2005).
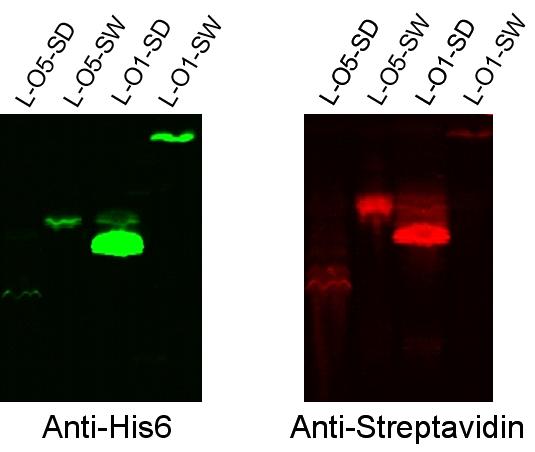
- Western blots were performed with transformed and induced cells, probing with anti-his6 antibody (each streptavidin had a His6 tag) and with anti-streptavidin antibody. Distinct bands were observed at the expected sizes in anti-his6 probing, and bands were observed at the same places with the anti-streptavidin probing. These results suggest the following.
- The promoter and ribosome binding site are functioning correctly for expression of these fusion protein construct.
- The BioBricks assembly of the fusion protein was successful in that reading frame was maintained, since the coding sequence for the His6 tag is found at the end of the construct.
- The streptavidin part of the fusion protein is still folding in such a way that anti-streptavidin antibody can recognize it.
Future Plans
- Now that we know that the construct is being expressed, we need to determine whether or not the construct is being localized to the outer membrane. We can separate the cell lysate by centrifugation, isolate the outer membrane proteins, and then perform Western blots, probing with anti-his6 and anti-streptavidin antibody.
- If the construct is indeed being localized to the outer membrane, we then need to determine if the streptavidin is being displayed functionally on the cell surface. We can probe whole cells with anti-streptavidin antibodies or even fluorescently tagged streptavidin aptamers for in-cell westerns, or we can visualize surface binding of biotinylated, fluorescently tagged oligonucleotides under a microscope.
- If streptavidin is being expressed on the cell surface, we can switch in other engineered streptavidin clones and compare biotin binding. We can also try adding a length of amino acids between OmpA and streptavidin, which might give spatial flexibility to allow formation of tetramers on the cell surface or to allow the streptavidin to extend outside of any extracellular complexes.
Electronic Notebook
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Fusion_proteins openwetware page]
References
- Francisco JA, Earhart CF, and Georgiou G. Transport and anchoring of beta-lactamase to the external surface of Escherichia coli. Proc Natl Acad Sci U S A 1992 Apr 1; 89(7) 2713-7.
- Earhart CF. Use of an Lpp-OmpA fusion vehicle for bacterial surface display. Methods Enzymol 2000; 326 506-16.
- Aslan FM, Yu Y, Mohr SC, and Cantor CR. Engineered single-chain dimeric streptavidins with an unexpected strong preference for biotin-4-fluorescein. Proc Natl Acad Sci U S A 2005 Jun 14; 102(24) 8507-12.
- Wu SC and Wong SL. Engineering soluble monomeric streptavidin with reversible biotin binding capability. J Biol Chem 2005 Jun 17; 280(24) 23225-31.
- Howarth M, Chinnapen DJ, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, El-Husseini A, and Ting AY. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods 2006 Apr; 3(4) 267-73.
- Phillips I, Silver P, A New Biobrick Assembly Strategy Designed for Facile Protein Engineering, MIT Synthetic Biology Working Group Reports, published online 20 April 2006 at MIT dspace.
A Circadian Oscillator for E.coli
Project Overview
- Oscillators are used in a variety of electronic circuits and would be essential in complex, time-dependant biocircuitry. The synthetic "repressilator" system engineered by Elowitz et al. (2000), though a good proof of concept, lacks the stability needed for long-term usage.
- Cyanobacteria have a naturally-evolved oscillator driven by the interactions between three proteins named KaiA, KaiB, and KaiC (see diagram at left). This "Kai clock" does not require transcriptional regulation and has been found to oscillate in vitro (Nakajima et al. 2005). It is also robust and can be modified for longer and shorter periods by point mutations in the Kai genes (Kondo et al. 1994).
- Based on these discoveries, our goal for the summer was to reconstitute the Kai oscillation cycle in E. coli. This would be the first step in creating a new oscillator part for the registry, as well as providing an important breakthrough for circadian biology research.
- The transcription factors that link the Kai clock to gene regulation in cyanobacteria are not well understood. Therefore we tested our clock in E. coli by measuring the amounts of phosphorylated and unphosphorylated KaiC via Western blot.
Results
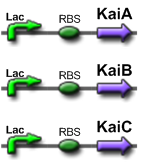
- We have succesfully created BioBricks of the Kai genes and several functional constructs. These include:
- Standalone KaiA , KaiB , and KaiC as Biobricks.(BBa_J36801, BBa_J36802, BBa_J36804)
- Kai proteins with BBa_J04500, which consists of a Lac promoter + RBS. (BBa_J36831, BBa_J36832, BBa_J36834)
- KaiA/J04500 + KaiC/J04500 in order to test KaiA's effect on KaiC (which, according to the model, should be promotion of KaiC's phosphorylation). (BBa_J36335)
- KaiB/J04500 + KaiC/J04500 in order to test KaiB's effect on KaiC (which, according to the model, should be nothing). (BBa_J36336)
- Based on the Tomita et al. 2005 model, we were able to develop hypotheses to predict the relative levels of phosphorylated/dephosphorylated KaiC in the presence of either KaiA or KaiB alone. Using the constructs we created, we tested and confirm these hypotheses. Anti-KaiC antibodies revealed a change in the relative concentration of dephosphorylated and phosphorylated KaiC from our different constructs.
- Hypotheses:
- KaiC alone will produce phosphorylated and dephosphorylated KaiC. This is based on the theory that KaiC autophosphorylates and autodephosphorylates.
- KaiA and KaiC will produce more phosphorylated than dephosphorylated KaiC, since KaiA promotes KaiC phosphorylation.
- KaiB will have no affect on KaiC's phosphorylation, since it does not interact with it directly according to the model. Therefore, the relative amounts of phosphorylated and dephosphorylated KaiC should be the same as when KaiC is expressed by itself.
- Western Blot results
- We performed a Western blot on our KaiC/J04500, KaiB/J04500 + KaiC/J04500, and KaiA/J04500 + KaiC/J04500 constructs using KaiC antibodies. Each sample was taken at the same OD. The results confirmed all of our hypotheses.
- In Lane 1 (KaiC/J04500), we can see a strong band of dephosphorylated KaiC and a weaker band of phosphorylated KaiC. This confirms that KaiC autophosphorylates and autodephosphorylates.
- In Lane 2(KaiB/J04500 + KaiC/J04500), we can again see a strong band of dephosphorylated KaiC and a weaker band of phosphorylated KaiC, in fact very similar, if not identical, to the results in Lane 1. This confirms that KaiB has no direct affect on KaiC's autophosphorylation, as predicted by the model.
- In Lane 3(KaiA/J04500 + KaiC/J04500), we see a strong band of dephosphorylated KaiC, but also a strong band of phosphorylated KaiC. Since the amount of phosphorylated KaiC is greater than that shown in Lanes 1 and 2, we have evidence that KaiA is interacting with KaiC to produce a greater amount of phosphorylated KaiC.
- in summary, we were able to express the three Kai proteins in E. coli, and show the predicted interaction between KaiA and KaiC and possible interaction between KaiB and KaiC.
Future Plans
- Short term plans include replicating our previous Western blot results as well as further characterizing post-transcriptional interactions between KaiA-KaiC and KaiB-KaiC pairs.
- In cyanobacteria, the Kai clock is reset by light/dark transitions. This serves to synchronize a population's circadian rhythms with the day/night cycle. This synchronization is necissary for experimenters to detect KaiC oscillation; a mixed-phase population will show no bulk, time-dependant change in KaiC phosphorylation state. Unfortunately, the molecular mechanism behind light/dark synchronization is not yet well understood, so we cannot transfer it to E.coli. Instead, we have designed a set of experiments using inducible promoters for the Kai genes to synchronize the Kai clocks within each cell and between cells in a culture . Our goal is to coerce a culture of E.coli cells into synchronized oscillation, which we can then measure by Western blot.
- As more is discovered about the intermediate factors between the Kai clock and gene regulation in cyanobacteria, we would like to leverage these findings to make the Kai clock drive other components in a biological circuit.
Electronic Notebook
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Cyanobacteria openwetware page]
Complete Electronic Notebooks
Detailed records of our summer activities, including results, can be found on openwetware, which the team used to host its [http://openwetware.org/wiki/IGEM:Harvard/2006 electronic notebook]. Links to specific project pages can be found in the above descriptions.