Imperial College 2006/Sandbox
From 2006.igem.org
| (17 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
|- | |- | ||
|width="20%" style="background:#2171B8"|[[Image:LogoFinal2.png|210px|center]] | |width="20%" style="background:#2171B8"|[[Image:LogoFinal2.png|210px|center]] | ||
| - | |width="40%" style="background:#2171B8" | <font face="arial" font style="color: #FFFFFF" > <center><u>'''Project Summary | + | |width="40%" style="background:#2171B8"| <font face="arial" font style="color: #FFFFFF" > <center><big><u>'''Project Summary'''</u></big></center> <br> Oscillators are a fundamental building block in many fields of engineering and are a widespread phenomenon in biology. Building a biological oscillator is thus a critical step forward in the field of Synthetic Biology. <br><br> |
| - | Oscillators are a fundamental building block in many fields of engineering and are a widespread phenomenon in biology. Building a biological oscillator is thus a critical step forward in the field of Synthetic Biology. <br><br> | + | |
'''Engineering a Molecular Predation Oscillator''', the iGEM project 2006 of Imperial College London, provides a new approach to creating a stable biological oscillator: It follows an engineering-based cycle of specification, design, modelling, implementation and testing/validation. The innovative design of the oscillator relies on predator-prey dynamics based on the Lotka-Volterra model. </font> | '''Engineering a Molecular Predation Oscillator''', the iGEM project 2006 of Imperial College London, provides a new approach to creating a stable biological oscillator: It follows an engineering-based cycle of specification, design, modelling, implementation and testing/validation. The innovative design of the oscillator relies on predator-prey dynamics based on the Lotka-Volterra model. </font> | ||
| - | |width="40%" style="background:#2171B8" | <font face="arial" font style="color: #FFFFFF" > <center><u>'''Achievements | + | |width="40%" style="background:#2171B8" | <font face="arial" font style="color: #FFFFFF" > <center><big><u>'''Achievements'''</u></big></center> <br> |
Following the design of the oscillator, a full theoretical analysis and detailed computer modelling of the Lotka-Volterra dynamics was carried out. These studies showed that it is theoretically possible to provide a stable oscillator. Because all components as well as the overall oscillator were modelled, its behaviour could be accurately predicted. Our team successfully built the functional parts, thus providing the building blocks for the final oscillator. All parts created were experimentally tested and their characterization could be used to feedback information into the modelling. | Following the design of the oscillator, a full theoretical analysis and detailed computer modelling of the Lotka-Volterra dynamics was carried out. These studies showed that it is theoretically possible to provide a stable oscillator. Because all components as well as the overall oscillator were modelled, its behaviour could be accurately predicted. Our team successfully built the functional parts, thus providing the building blocks for the final oscillator. All parts created were experimentally tested and their characterization could be used to feedback information into the modelling. | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
</font> | </font> | ||
|} | |} | ||
| Line 24: | Line 26: | ||
| width="20%" style="background:#2171B8"| <center>[[Image:Oscillator Icon.PNG|100px|center]] </center> | | width="20%" style="background:#2171B8"| <center>[[Image:Oscillator Icon.PNG|100px|center]] </center> | ||
| style="background:#2171B8" colspan="3"| <font face="arial" font style="color: #FFFFFF"> <center><big>'''Engineering a Molecular Predation Oscillator'''</big><br> | | style="background:#2171B8" colspan="3"| <font face="arial" font style="color: #FFFFFF"> <center><big>'''Engineering a Molecular Predation Oscillator'''</big><br> | ||
| - | For building our biological oscillator, the engineering cycle below was followed.<br> '''Click on each of the stages of the engineering cycle below in order to find out about the different stages & aspects of the project. '''</center | + | For building our biological oscillator, the engineering cycle below was followed.<br> |
| + | '''Click on each of the stages of the engineering cycle below in order to find out about the different stages & aspects of the project. '''</center> | ||
|- | |- | ||
| style="background:#2171B8" width="200pt" rowspan="4" | | | style="background:#2171B8" width="200pt" rowspan="4" | | ||
| Line 34: | Line 37: | ||
*Mimics predator-prey dynamics based on the Lotka-Volterra model | *Mimics predator-prey dynamics based on the Lotka-Volterra model | ||
'''3) Modelling: ''' | '''3) Modelling: ''' | ||
| - | *Full theoretical analysis | + | *Full theoretical analysis [http://www.openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/Theoretical_Analyses] |
*Modelling of the full system | *Modelling of the full system | ||
'''4) Implementation: ''' | '''4) Implementation: ''' | ||
| Line 45: | Line 48: | ||
| align="left"|[[Image:ArrowCurvedDown.PNG|120px]] | | align="left"|[[Image:ArrowCurvedDown.PNG|120px]] | ||
|- | |- | ||
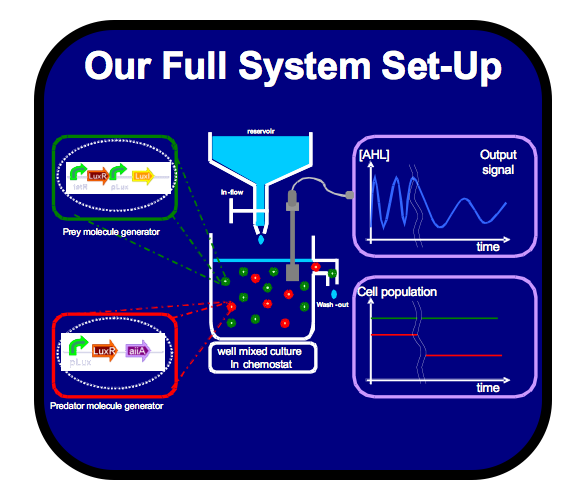
| - | | align="center"|[http://www.openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System | + | | align="center"|[http://www.openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System http://openwetware.org/images/f/fd/IGEM_IMPERIAL_DevCycle_Specifications.png] |
| align="center" rowspan="2"|[[Image:IGEM_IMPERIAL_DevCycle_FullSystem.png |206px|center]] | | align="center" rowspan="2"|[[Image:IGEM_IMPERIAL_DevCycle_FullSystem.png |206px|center]] | ||
| align="center"|[http://www.openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System/Modelling http://openwetware.org/images/5/5f/IGEM_IMPERIAL_DevCycle_Modelling.png] | | align="center"|[http://www.openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System/Modelling http://openwetware.org/images/5/5f/IGEM_IMPERIAL_DevCycle_Modelling.png] | ||
| Line 52: | Line 55: | ||
| align="center"|[[Image:ArrowDown.png|20px]] | | align="center"|[[Image:ArrowDown.png|20px]] | ||
|- | |- | ||
| - | | align="center"|[http://www.openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System/ | + | | align="center"|[http://www.openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System/TestingValidation http://openwetware.org/images/2/20/IGEM_IMPERIAL_DevCycle_Validation.png] |
| align="center"|[[Image:ArrowLeft.png|150px]] | | align="center"|[[Image:ArrowLeft.png|150px]] | ||
| align="center"|[http://www.openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System/Implementation http://openwetware.org/images/0/04/IGEM_IMPERIAL_DevCycle_Implementation.png] | | align="center"|[http://www.openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System/Implementation http://openwetware.org/images/0/04/IGEM_IMPERIAL_DevCycle_Implementation.png] | ||
| Line 66: | Line 69: | ||
*This part is placed downstream of a promoter and prevents any Pops from the promoter passing through this part | *This part is placed downstream of a promoter and prevents any Pops from the promoter passing through this part | ||
*When an accompanying Cre Recombinase plasmid becomes activated, the enzyme produced will permanently cut a section of DNA from the plasmid containing this part | *When an accompanying Cre Recombinase plasmid becomes activated, the enzyme produced will permanently cut a section of DNA from the plasmid containing this part | ||
| - | *Only then, the polymerase can pass through this part and transcribe downstream genes. </font> | + | *Only then, the polymerase can pass through this part and transcribe downstream genes. |
| - | | style="background:#2171B8" width="50%"| | + | </font> |
| + | | style="background:#2171B8" width="50%" | | ||
<font face="arial" font style="color: #FFFFFF">We also worked on a Biosensor for measuring AHL concentrations in order to establish a Biological to Electrical Interface this summer. | <font face="arial" font style="color: #FFFFFF">We also worked on a Biosensor for measuring AHL concentrations in order to establish a Biological to Electrical Interface this summer. | ||
*Using an enzyme to hydrolize the lactone AHL would result in a local change in pH | *Using an enzyme to hydrolize the lactone AHL would result in a local change in pH | ||
| - | *Measuring the change in pH gives a measurement of how much AHL is present.</font> | + | *Measuring the change in pH gives a measurement of how much AHL is present. |
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | </font> | ||
|} | |} | ||
| Line 122: | Line 131: | ||
==Our Open Documentation== | ==Our Open Documentation== | ||
| + | |||
| + | *Full Documentation, 100's of Photographs, Extensive Modelling, Lab Notebook and much more! | ||
| + | |||
__NOTOC__ | __NOTOC__ | ||
{| class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 5px; background-color: #ffffff; border: 2px solid #2171B8;text-align:center" | {| class="wikitable" border="0" cellpadding="10" cellspacing="1" style="padding: 5px; background-color: #ffffff; border: 2px solid #2171B8;text-align:center" | ||
| Line 141: | Line 153: | ||
*[http://openwetware.org/wiki/IGEM:IMPERIAL/2006/Photos <font face="arial" font style="color: #FFFFFF">Photos</font>] | *[http://openwetware.org/wiki/IGEM:IMPERIAL/2006/Photos <font face="arial" font style="color: #FFFFFF">Photos</font>] | ||
|} | |} | ||
| - | |||
==The Team and Acknowledgements== | ==The Team and Acknowledgements== | ||
| Line 172: | Line 183: | ||
*[http://www3.imperial.ac.uk/people/p.freemont <font face="arial" font style="color: #FFFFFF">Prof. Paul Freemont</font>] | *[http://www3.imperial.ac.uk/people/p.freemont <font face="arial" font style="color: #FFFFFF">Prof. Paul Freemont</font>] | ||
*[http://www3.imperial.ac.uk/people/d.mann <font face="arial" font style="color: #FFFFFF">Dr. David Mann</font>] | *[http://www3.imperial.ac.uk/people/d.mann <font face="arial" font style="color: #FFFFFF">Dr. David Mann</font>] | ||
| - | *<font face=" | + | *<font face="arial" font style="color: #FFFFFF">Kirsten Jensen</font> |
*[http://openwetware.org/wiki/User:Vincent <font face="arial" font style="color: #FFFFFF">Vincent Rouilly</font>] | *[http://openwetware.org/wiki/User:Vincent <font face="arial" font style="color: #FFFFFF">Vincent Rouilly</font>] | ||
*[http://openwetware.org/wiki/User:chuehloo <font face="arial" font style="color: #FFFFFF">Chueh-Loo Poh</font>] | *[http://openwetware.org/wiki/User:chuehloo <font face="arial" font style="color: #FFFFFF">Chueh-Loo Poh</font>] | ||
| Line 180: | Line 191: | ||
<center> | <center> | ||
| + | |||
==<font face="arial" font style="color: #FFFFFF">Funding</font>== | ==<font face="arial" font style="color: #FFFFFF">Funding</font>== | ||
</center> | </center> | ||
Latest revision as of 12:57, 30 October 2006
| Oscillators are a fundamental building block in many fields of engineering and are a widespread phenomenon in biology. Building a biological oscillator is thus a critical step forward in the field of Synthetic Biology. Engineering a Molecular Predation Oscillator, the iGEM project 2006 of Imperial College London, provides a new approach to creating a stable biological oscillator: It follows an engineering-based cycle of specification, design, modelling, implementation and testing/validation. The innovative design of the oscillator relies on predator-prey dynamics based on the Lotka-Volterra model. | Following the design of the oscillator, a full theoretical analysis and detailed computer modelling of the Lotka-Volterra dynamics was carried out. These studies showed that it is theoretically possible to provide a stable oscillator. Because all components as well as the overall oscillator were modelled, its behaviour could be accurately predicted. Our team successfully built the functional parts, thus providing the building blocks for the final oscillator. All parts created were experimentally tested and their characterization could be used to feedback information into the modelling.
|
Main Project
Secondary Projects
| [http://openwetware.org/wiki/IGEM:IMPERIAL/2006/project/popsblocker PoPs Blocker] | [http://openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Bio_elec_interface Biological to Electrical Interface] |
|---|---|
|
As a method of controlling the activation of the positive-feedback loop in our predator-prey based oscillator, we successfully created this part, which can be used as a general Pops Blocker:
|
We also worked on a Biosensor for measuring AHL concentrations in order to establish a Biological to Electrical Interface this summer.
|
Our Contributions to the Registry
| Part Logo | Description | Link to registry | Built | Tested | Characterized | Sequenced | Sent |
|---|---|---|---|---|---|---|---|
| Final Constructs | |||||||
| Final Prey Cell | J37015 | YES | YES | Pending | YES | YES | |
| | Cre/Lox Prey Control | J37027 | YES | YES | YES | YES | YES |
| Test Constructs | |||||||
| Final Polycistronic Predator Cell Test Construct | J37016 | YES | YES | YES | YES | YES | |
| | Predator Cell, pLux Transfer Function (two promoters) | J37020 | YES | YES | NO | NO | YES |
| Intermediate Parts | |||||||
| | AHL induced LuxR generator (for predator cell) | J37019 | YES | N/A | N/A | YES | YES |
| | RBS + LuxR | J37033 | YES | N/A | N/A | YES | YES |
| | Prey Cell Intermediate | J37034 | YES | N/A | N/A | YES | YES |
| | LuxR + GFP | J37032 | YES | N/A | N/A | YES | YES |
Our Open Documentation
- Full Documentation, 100's of Photographs, Extensive Modelling, Lab Notebook and much more!
|
|
|
|
|
|---|
The Team and Acknowledgements
Undergrads
|
Advisors
|
Funding
|
|---|