Ljubljana, Slovenia 2006/Results & Conclusions
From 2006.igem.org
Jelka.pohar (Talk | contribs) |
|||
| (66 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<table cellpadding="10"> | <table cellpadding="10"> | ||
<tr><th>[[Ljubljana, Slovenia 2006|Home]]</th> | <tr><th>[[Ljubljana, Slovenia 2006|Home]]</th> | ||
| - | <th>[[Ljubljana, Slovenia 2006/Background and Signalling Pathway|Background and | + | <th>[[Ljubljana, Slovenia 2006/Background and Signalling Pathway|Background and Signaling Pathway]]</th> |
<th>[[Ljubljana, Slovenia 2006/Project & Model|Project & Model]]</th> | <th>[[Ljubljana, Slovenia 2006/Project & Model|Project & Model]]</th> | ||
<th>[[Ljubljana, Slovenia 2006/Methods|Methods]]</th> | <th>[[Ljubljana, Slovenia 2006/Methods|Methods]]</th> | ||
| Line 18: | Line 18: | ||
We prepared individual BioBricks that could be used in mammalian cells. The following BioBricks were used: | We prepared individual BioBricks that could be used in mammalian cells. The following BioBricks were used: | ||
*promoter (constitutive CMV and [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] inducible promoter), | *promoter (constitutive CMV and [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] inducible promoter), | ||
| - | *terminator (which we included into plasmids to avoid repeated additional steps in construction of each functional Part | + | *terminator (which we included into plasmids to avoid repeated additional steps in construction of each functional Part) |
*protein coding sequences, which include: | *protein coding sequences, which include: | ||
**two inhibitory (dominant negative) proteins of the signaling cascade ([[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] and dn[[Ljubljana, Slovenia 2006/Terms & References#Terms|TRAF6]]), | **two inhibitory (dominant negative) proteins of the signaling cascade ([[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] and dn[[Ljubljana, Slovenia 2006/Terms & References#Terms|TRAF6]]), | ||
| Line 90: | Line 90: | ||
<br> | <br> | ||
| - | <h2> | + | <h2>Detection of phosphorylation of ERK kinases by flow cytometry reflects the activation of the TLR signaling </h2> |
| - | < | + | <br style="clear:both;"/> |
| + | [[Image:cito1.gif|cito1.gif|center|thumb|400px|<b> Figure 19: Flow cytometric profile of phosphorylated ERK kinases in HEK293 cells stimulated by LPS for 0, 20 and 80 minutes.</b>]] | ||
| - | < | + | <br style="clear:both;"/> |
| - | [[Image: | + | |
| + | Flow Cytometry experiments showed that after 20 minutes after the second stimulation (80 min after the first stimulation) the increase in the level of phosphorylated ERK kinases was evident(Figure 19). The method was not as sensitive to the rapid response as we hoped. Further optimization of the method would be necessary in order to obtain the quantitative data to accurately model the cell signaling. | ||
| + | |||
| + | <h2>Detection of transcriptionally competent [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] in whole cell extracts dependent on the time after stimulation with [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]] determined with promoter ELISA method</h2> | ||
| + | |||
| + | <br style="clear:both;"/> | ||
| + | |||
| + | [[Image:Elisa_results.gif|center|thumb|800px|<b>Figure 20: Activity of peroxidase (conjugated on secondary antibodies) in luminescence units as a function of time after stimulation with [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]].</b> The amount of free [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] (and consequently luminescence) increases with time of stimulation (7 hours) and decreases after extended (23 hours) stimulation.]] | ||
| + | |||
| + | <br style="clear:both;"/> | ||
| + | |||
| + | Cells were transfected with [[Ljubljana, Slovenia 2006/Terms & References#Terms|TLR4]] plasmid and [[Ljubljana, Slovenia 2006/Terms & References#Terms|MD-2]] plasmid in order to confer the LPS responsiveness. We have shown that cells with TRL4 receptor respond to [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]] stimulation and that ELISA is apropriate and sensitive method for the detection of stimulation. However we did not get satisfying results when stimulating (with [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]] or flagellin) cells transfected neither with [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] nor [[Ljubljana, Slovenia 2006/Terms & References#Terms|TRAF6]]. The response of the ELISA method, as the most direct measurement of the free NF-κB was not sufficiently reproducible and linear in comparison to the method of luciferase reporter plasmids (see below). | ||
| + | |||
| + | <h2>Inducible transcription by the stimulation of Toll-like receptor</h2> | ||
| + | |||
| + | <p>Stimulation of [[Ljubljana, Slovenia 2006/Terms & References#Terms|HEK293]] cells transfected by TLR with their agonist (flagellin in case of TLR5) stimulates the translocation of [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] into the nucleus and activation of transcription of [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]]-responsive genes. We have used the firefly luciferase under the [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] dependent promoter to monitor the promotor activation. We tested the functionality of our part containing [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] dependent promotor in a construct with [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]]-rLuc. We detected the R-luciferase activity only in stimulated cells with significant activity appearing a few hours after the cell stimulation, which proves the functionality of this part (inducible promoter).</p> | ||
| + | |||
| + | <br style="clear:both;"/> | ||
| + | |||
| + | |||
| + | [[Image:graf_gabi.gif|left|thumb|400px|<b>Figure 21: Transcription of [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] with rLuc reporter is induced by bacterial flagellin </b> Activity of ''Renilla'' luciferase stimulated by the addition of a pulse of [[Ljubljana, Slovenia 2006/Terms & References#Terms|flagellin]]. [[Ljubljana, Slovenia 2006/Terms & References#Terms|HEK293]] cells were transfected with device comprising [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]]-inducible fusion of [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]]-rLuc.]] | ||
| + | |||
| + | [[Image:rezultati7.gif|left|thumb|400px|<b>Figure 22: Transcription of [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] with rLuc reporter in our device is inducible by bacterial flagellin.</b> Activity of the Firefly luciferase stimulated by the addition of two pulses of [[Ljubljana, Slovenia 2006/Terms & References#Terms|flagellin]]. ]] | ||
| + | |||
| + | <br style="clear:both;"/> | ||
| + | <p>We also measured NF-kB-responsive F-luciferase activity of this device. We expected a decrease in the activity after the amount of time sufficient for the sysnthesis of inhibitory fusion protein in cells. To our dissapointment no decrease of the cell activation was observed. One of the possible causes for the lack of inhibition of the dnMyD88-rLuc protein fusion could be the sterical hindrance of the activity of dnMyD88. This domain interacts with TIR domain of TLRs in the direct proximity of the cellular membrane, which could prevent interaction with larger protein domains. Based on those results we also conducted our experiments with a device containing dnMyD88 without of any protein appendix. | ||
| - | <h2>Inhibition of cell signaling by a feedback device</h2> | + | <h2>Inhibition of cell signaling by a dnMyD88 feedback device</h2> |
| - | + | An adapter protein [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]], consisting of a TIR and death domain, is at the crossroads of Toll-like receptors. Therefore activation of each of the surface expressed TLR recruits [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] to the cell membrane. We selected to use the receptor TLR5 and flagellin instead of the often used [[Ljubljana, Slovenia 2006/Terms & References#Terms|TLR4]] and its agonist [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]], because the [[Ljubljana, Slovenia 2006/Terms & References#Terms|TLR4]] system is extremely sensitive to the contamination with bacterial [[Ljubljana, Slovenia 2006/Terms & References#Terms|LPS]], which often copurifies with the plasmid. In this way we have avoided the contamination issue, but our system is designed to work on any signaling receptor that has [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] in its signaling pathway (i.e. TLR1,2,4,5,6,7,8,9,11,IL-1). | |
Cells were cotransfected with TLR5, reporter plasmid (NFκB-Fluc) and our feedback device ([[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]]-[[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]]).</p> | Cells were cotransfected with TLR5, reporter plasmid (NFκB-Fluc) and our feedback device ([[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]]-[[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]]).</p> | ||
| - | <p>We expect that after stimulation with flagellin cytoplasmic [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] is released from its inhibitor. It then migrates into the nucleus and induces the transcription of our construct. Accumulation of the dominant negative protein should compete with wild-type [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] and attenuate the downstream | + | <p>We expect that after stimulation with flagellin cytoplasmic [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] is released from its inhibitor. It then migrates into the nucleus and induces the transcription of our construct. Accumulation of the dominant negative protein should compete with wild-type [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] and attenuate the downstream signaling pathway. We can detect the [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] translocation, transcriptional activation and phosphorylation by our detection systems - luminescence, free [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]], phosphorylation detected by flow cytometry and confocal microscopy. In case that our construct is followed by [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]] (degradation) sequence, it is expected that product should degrade rapidly and shorten the inhibition period. If the stimulus is still present, cells should again respond to it and the cycle with inhibition will repeat as well.</p> |
<p>We have designed the experiment by two consecutive pulses of TLR stimulation separated by 4-6 hours, which is sufficient time to accumulate the inducible inhibitor. Normal cells should respond to both stimuli, while the cells with our device should respond to the same extent only to the first pulse -- this induces the transcription of the inhibitor, therefore the response to the second stimulus should be decreased. However if the stimuli are separated by a longer timespan, the system should respond normally, as the inhibitor has already degraded and the systemn has reset to the normal state.</p> | <p>We have designed the experiment by two consecutive pulses of TLR stimulation separated by 4-6 hours, which is sufficient time to accumulate the inducible inhibitor. Normal cells should respond to both stimuli, while the cells with our device should respond to the same extent only to the first pulse -- this induces the transcription of the inhibitor, therefore the response to the second stimulus should be decreased. However if the stimuli are separated by a longer timespan, the system should respond normally, as the inhibitor has already degraded and the systemn has reset to the normal state.</p> | ||
<center> | <center> | ||
| - | [[Image:rezultati1a. | + | [[Image:rezultati1a.gif|center|thumb|400px|<b>Figure 23: [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] feedback loop decreases the cellular activation by repeated stimulation with bacterial [[Ljubljana, Slovenia 2006/Terms & References#Terms|flagellin]]. </b> Normalized luciferase activity (Fluc/Rluc) of cells stimulated by the addition of two pulses of flagellin separated by 6 hours, indicated time after the second pulse is shown. [[Ljubljana, Slovenia 2006/Terms & References#Terms|HEK293]] cells were transfected with device comprising NFκB-inducible [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] in comparison to wild type.]] |
</center> | </center> | ||
| + | <br style="clear:both;"/> | ||
<p>Our results show that the system with inducible [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] indeed responds weaker to the second stimulus. The minimal delay between two stimuli should correspond to the time required for the synthesis of the inhibitor, which is exactly as planned, since it does not completely abolish the cellular response but decreases it after extended stimulation.</p> | <p>Our results show that the system with inducible [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] indeed responds weaker to the second stimulus. The minimal delay between two stimuli should correspond to the time required for the synthesis of the inhibitor, which is exactly as planned, since it does not completely abolish the cellular response but decreases it after extended stimulation.</p> | ||
| + | |||
| + | <br style="clear:both;"/> | ||
<h2>Decrease of the protein lifetime by the [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST sequence]]</h2> | <h2>Decrease of the protein lifetime by the [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST sequence]]</h2> | ||
| Line 117: | Line 146: | ||
<p>Rapid degradation signature motif - "[[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]]" was considered to tune the lifetime of the inhibitor in order to reset the cells to the normal responsiveness more rapidly. Fusion of the luciferase and [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST sequence]] under the control of CMV (Device [http://partsregistry.org/Part:BBa_J52039 BBa_J52039]) was used to test the effectiveness of the [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST sequence]]. We have used the cycloheximide to stop the protein synthesis and analyse the lifetime of the fusion protein with [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]] sequence. The steady state of the luciferase activity was 5-10 fold lower in the Rluc-[[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]] fusin than without of the [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]] signature, as evident from the chart below.</p> | <p>Rapid degradation signature motif - "[[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]]" was considered to tune the lifetime of the inhibitor in order to reset the cells to the normal responsiveness more rapidly. Fusion of the luciferase and [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST sequence]] under the control of CMV (Device [http://partsregistry.org/Part:BBa_J52039 BBa_J52039]) was used to test the effectiveness of the [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST sequence]]. We have used the cycloheximide to stop the protein synthesis and analyse the lifetime of the fusion protein with [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]] sequence. The steady state of the luciferase activity was 5-10 fold lower in the Rluc-[[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]] fusin than without of the [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST]] signature, as evident from the chart below.</p> | ||
| - | [[Image:graf1.gif|thumb|center|800px| | + | [[Image:graf1.gif|thumb|center|800px|<b>Figure 24: Addition of the PEST degradation tag decreases the steady state of the protein in cells due to its rapid degradation </b>. Cycloheximide was added to cells in order to stop the protein synthesis and monitor the protein degradation kinetics.]] |
<p>Constructs of the [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] domain and [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST sequence]] were not effective as inhibitors of signaling because of the interference of the C-terminal extension of the [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] inhibitory domain, as described above for the fusion constructs with the reporter luciferase or GFP.</p> | <p>Constructs of the [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] domain and [[Ljubljana, Slovenia 2006/Terms & References#Terms|PEST sequence]] were not effective as inhibitors of signaling because of the interference of the C-terminal extension of the [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] inhibitory domain, as described above for the fusion constructs with the reporter luciferase or GFP.</p> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<div style="float:left; background-color:#cedff2; padding: .2em .6em; border: 1px solid black">''' | <div style="float:left; background-color:#cedff2; padding: .2em .6em; border: 1px solid black">''' | ||
| Line 132: | Line 155: | ||
<h1>Conclusions</h1> | <h1>Conclusions</h1> | ||
| - | *the same principles of BioBricks can be used in the mammalian cell system as in bacteria and | + | *the same principles of BioBricks can be used in the mammalian cell system as in bacteria and yeasts |
*we have succesfully implemented a feedback loop that decreases the cellular activation with some delay | *we have succesfully implemented a feedback loop that decreases the cellular activation with some delay | ||
*response of this feedback loop is transient and the cell responsiveness is restored after the synthesized inhibitor has degraded | *response of this feedback loop is transient and the cell responsiveness is restored after the synthesized inhibitor has degraded | ||
*[[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] inhibitor was not effective as a C-terminal fusion with luciferase or GFP, which may be due to the steric hindrance; it is likely that N-terminal fusion of reporter domains would be functional as signaling inhibitors | *[[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] inhibitor was not effective as a C-terminal fusion with luciferase or GFP, which may be due to the steric hindrance; it is likely that N-terminal fusion of reporter domains would be functional as signaling inhibitors | ||
| - | *our constructed device mimicks the natural mechanism of tolerance only that it is activated faster, which may | + | *our constructed device mimicks the natural mechanism of tolerance only that it is activated faster, which may benefitial for an organism, |
*simplified model of the TLR signaling qualitatively captures most of the features of the natural system | *simplified model of the TLR signaling qualitatively captures most of the features of the natural system | ||
</div> | </div> | ||
| Line 142: | Line 165: | ||
| + | |||
| + | <br> | ||
<h2>Suggestions</h2> | <h2>Suggestions</h2> | ||
| Line 150: | Line 175: | ||
<table cellpadding="10"> | <table cellpadding="10"> | ||
<tr><th>[[Ljubljana, Slovenia 2006|Home]]</th> | <tr><th>[[Ljubljana, Slovenia 2006|Home]]</th> | ||
| - | <th>[[Ljubljana, Slovenia 2006/Background and Signalling Pathway|Background and | + | <th>[[Ljubljana, Slovenia 2006/Background and Signalling Pathway|Background and Signaling Pathway]]</th> |
<th>[[Ljubljana, Slovenia 2006/Project & Model|Project & Model]]</th> | <th>[[Ljubljana, Slovenia 2006/Project & Model|Project & Model]]</th> | ||
<th>[[Ljubljana, Slovenia 2006/Methods|Methods]]</th> | <th>[[Ljubljana, Slovenia 2006/Methods|Methods]]</th> | ||
Latest revision as of 13:25, 2 April 2007
| Home | Background and Signaling Pathway | Project & Model | Methods | Terms & References | Team members |
|---|
Results
Construction of Biobricks
We prepared individual BioBricks that could be used in mammalian cells. The following BioBricks were used:
- promoter (constitutive CMV and NF-κB inducible promoter),
- terminator (which we included into plasmids to avoid repeated additional steps in construction of each functional Part)
- protein coding sequences, which include:
- two inhibitory (dominant negative) proteins of the signaling cascade (dnMyD88 and dnTRAF6),
- two reporters: Renilla luciferase (in order to allow simultaneous dual luciferase assay to normalize the luminescence for the efficiency of transfection and cell number) and GFP
- PEST sequence to decrease the lifetime of the inhibitor.
Linkers between the protein coding sequences (e.g. MyD88+luciferase+PEST) were prepared by PCR ligation to avoid addition of unwanted aminoacids in the linker region between the protein domains.
| Registration number | Part's Name | Part/Device | Vector |
|---|---|---|---|
| [http://partsregistry.org/Part:BBa_J52008 BBa_J52008] | rluc | Part | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52010 BBa_J52010] | NFκB | Part | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52011 BBa_J52011] | dnMyD88-likn-rLuc | Part | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52012 BBa_J52012] | rluc-link-PEST191 | Part | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52013 BBa_J52013] | dnMyD88-link-rluc-link-pest191 | Part | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52014 BBa_J52014] | NFκB+dnMyD88-link-rLuc | Device | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52016 BBa_J52016] | eukaryotic terminator | Part | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52017 BBa_J52017] | eukaryotic terminator vector | pSB1AK3 | |
| [http://partsregistry.org/Part:BBa_J52018 BBa_J52018] | NFκB+rLuc | Device | pSB1AC3 |
| [http://partsregistry.org/Part:BBa_J52019 BBa_J52019] | dnTRAF6 | Part | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52021 BBa_J52021] | dnTRAF6-link-GFP | Part | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52022 BBa_J52022] | NFκB+dnTRAF6-link-GFP | Device | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52023 BBa_J52023] | NFκB+rLuc-link-PEST191 | Device | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52024 BBa_J52024] | NFκB+dnMyD88-link-rLuc-link-PEST191 | Device | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52026 BBa_J52026] | dnMyD88-link-GFP | Part | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52027 BBa_J52027] | NFκB+dnMyD88-link-GFP | Device | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52028 BBa_J52028] | GFP-PEST191 | Part | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52029 BBa_J52029] | NFκB+GFP-PEST191 | Device | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52034 BBa_J52034] | CMV | Part | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52035 BBa_J52035] | dnMyD88 | Part | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52036 BBa_J52036] | NFκB+dnMyD88 | Device | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52038 BBa_J52038] | CMV-rLuc | Device | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52039 BBa_J52039] | CMV+rLuc-link-PEST191 | Device | pSB1A2 |
| [http://partsregistry.org/Part:BBa_J52040 BBa_J52040] | CMV+GFP-PEST191 | Device | pSB1AK3 |
| [http://partsregistry.org/Part:BBa_J52642 BBa_J52642] | GFP | Part | pSB1AK3+TER |
| [http://partsregistry.org/Part:BBa_J52648 BBa_J52648] | CMV+GFP | Device | pSB1AK3+TER |
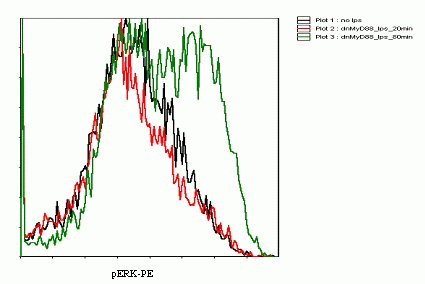
Detection of phosphorylation of ERK kinases by flow cytometry reflects the activation of the TLR signaling
Flow Cytometry experiments showed that after 20 minutes after the second stimulation (80 min after the first stimulation) the increase in the level of phosphorylated ERK kinases was evident(Figure 19). The method was not as sensitive to the rapid response as we hoped. Further optimization of the method would be necessary in order to obtain the quantitative data to accurately model the cell signaling.
Detection of transcriptionally competent NF-κB in whole cell extracts dependent on the time after stimulation with LPS determined with promoter ELISA method
Cells were transfected with TLR4 plasmid and MD-2 plasmid in order to confer the LPS responsiveness. We have shown that cells with TRL4 receptor respond to LPS stimulation and that ELISA is apropriate and sensitive method for the detection of stimulation. However we did not get satisfying results when stimulating (with LPS or flagellin) cells transfected neither with dnMyD88 nor TRAF6. The response of the ELISA method, as the most direct measurement of the free NF-κB was not sufficiently reproducible and linear in comparison to the method of luciferase reporter plasmids (see below).
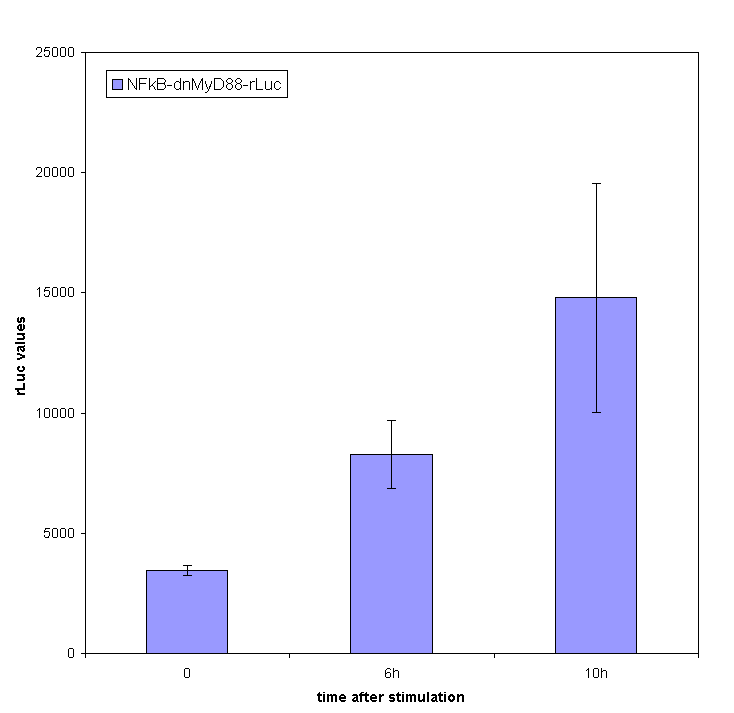
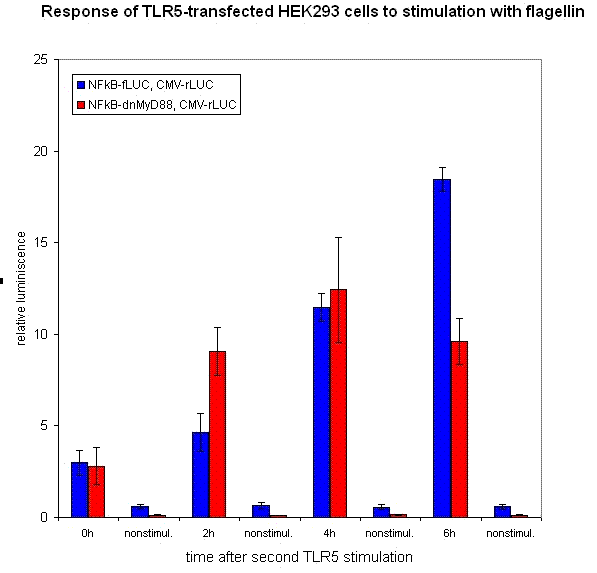
Inducible transcription by the stimulation of Toll-like receptor
Stimulation of HEK293 cells transfected by TLR with their agonist (flagellin in case of TLR5) stimulates the translocation of NF-κB into the nucleus and activation of transcription of NF-κB-responsive genes. We have used the firefly luciferase under the NF-κB dependent promoter to monitor the promotor activation. We tested the functionality of our part containing NF-κB dependent promotor in a construct with dnMyD88-rLuc. We detected the R-luciferase activity only in stimulated cells with significant activity appearing a few hours after the cell stimulation, which proves the functionality of this part (inducible promoter).
We also measured NF-kB-responsive F-luciferase activity of this device. We expected a decrease in the activity after the amount of time sufficient for the sysnthesis of inhibitory fusion protein in cells. To our dissapointment no decrease of the cell activation was observed. One of the possible causes for the lack of inhibition of the dnMyD88-rLuc protein fusion could be the sterical hindrance of the activity of dnMyD88. This domain interacts with TIR domain of TLRs in the direct proximity of the cellular membrane, which could prevent interaction with larger protein domains. Based on those results we also conducted our experiments with a device containing dnMyD88 without of any protein appendix.
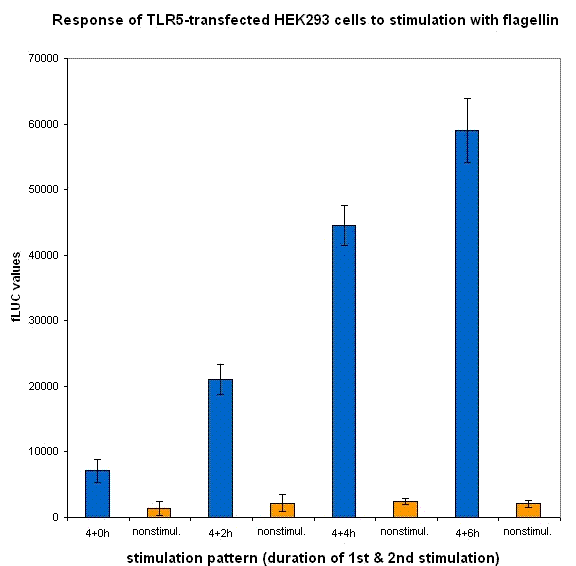
Inhibition of cell signaling by a dnMyD88 feedback device
An adapter protein MyD88, consisting of a TIR and death domain, is at the crossroads of Toll-like receptors. Therefore activation of each of the surface expressed TLR recruits MyD88 to the cell membrane. We selected to use the receptor TLR5 and flagellin instead of the often used TLR4 and its agonist LPS, because the TLR4 system is extremely sensitive to the contamination with bacterial LPS, which often copurifies with the plasmid. In this way we have avoided the contamination issue, but our system is designed to work on any signaling receptor that has MyD88 in its signaling pathway (i.e. TLR1,2,4,5,6,7,8,9,11,IL-1).
Cells were cotransfected with TLR5, reporter plasmid (NFκB-Fluc) and our feedback device (NF-κB-dnMyD88).We expect that after stimulation with flagellin cytoplasmic NF-κB is released from its inhibitor. It then migrates into the nucleus and induces the transcription of our construct. Accumulation of the dominant negative protein should compete with wild-type MyD88 and attenuate the downstream signaling pathway. We can detect the NF-κB translocation, transcriptional activation and phosphorylation by our detection systems - luminescence, free NF-κB, phosphorylation detected by flow cytometry and confocal microscopy. In case that our construct is followed by PEST (degradation) sequence, it is expected that product should degrade rapidly and shorten the inhibition period. If the stimulus is still present, cells should again respond to it and the cycle with inhibition will repeat as well.
We have designed the experiment by two consecutive pulses of TLR stimulation separated by 4-6 hours, which is sufficient time to accumulate the inducible inhibitor. Normal cells should respond to both stimuli, while the cells with our device should respond to the same extent only to the first pulse -- this induces the transcription of the inhibitor, therefore the response to the second stimulus should be decreased. However if the stimuli are separated by a longer timespan, the system should respond normally, as the inhibitor has already degraded and the systemn has reset to the normal state.
Our results show that the system with inducible dnMyD88 indeed responds weaker to the second stimulus. The minimal delay between two stimuli should correspond to the time required for the synthesis of the inhibitor, which is exactly as planned, since it does not completely abolish the cellular response but decreases it after extended stimulation.
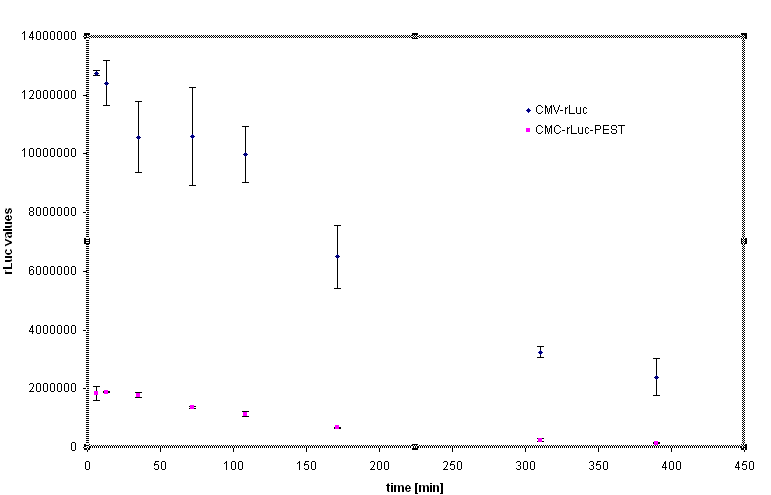
Decrease of the protein lifetime by the PEST sequence
Rapid degradation signature motif - "PEST" was considered to tune the lifetime of the inhibitor in order to reset the cells to the normal responsiveness more rapidly. Fusion of the luciferase and PEST sequence under the control of CMV (Device [http://partsregistry.org/Part:BBa_J52039 BBa_J52039]) was used to test the effectiveness of the PEST sequence. We have used the cycloheximide to stop the protein synthesis and analyse the lifetime of the fusion protein with PEST sequence. The steady state of the luciferase activity was 5-10 fold lower in the Rluc-PEST fusin than without of the PEST signature, as evident from the chart below.
Constructs of the dnMyD88 domain and PEST sequence were not effective as inhibitors of signaling because of the interference of the C-terminal extension of the MyD88 inhibitory domain, as described above for the fusion constructs with the reporter luciferase or GFP.
Conclusions
- the same principles of BioBricks can be used in the mammalian cell system as in bacteria and yeasts
- we have succesfully implemented a feedback loop that decreases the cellular activation with some delay
- response of this feedback loop is transient and the cell responsiveness is restored after the synthesized inhibitor has degraded
- dnMyD88 inhibitor was not effective as a C-terminal fusion with luciferase or GFP, which may be due to the steric hindrance; it is likely that N-terminal fusion of reporter domains would be functional as signaling inhibitors
- our constructed device mimicks the natural mechanism of tolerance only that it is activated faster, which may benefitial for an organism,
- simplified model of the TLR signaling qualitatively captures most of the features of the natural system
Suggestions
- for mammalian cell systems an additional set of vectors may be constructed based on the backbone, which includes the selection marker for stable transfection or retroviral integration
| Home | Background and Signaling Pathway | Project & Model | Methods | Terms & References | Team members |
|---|