Next steps
From 2006.igem.org
m (updated partinfo tag of pSB1A7) |
|||
| (2 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
[[Image:dc_control.jpg]] | [[Image:dc_control.jpg]] | ||
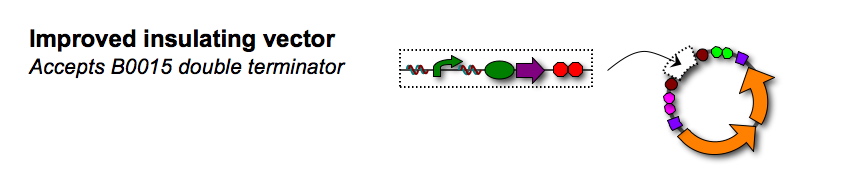
| - | Our new vector pSB1A7 eliminates the read-through problems we had with <partinfo>pSB1A3</partinfo>, but is incompatible with double terminator <partinfo>B0015</partinfo>. We hope to design and build a new vector that is well-insulated from read-through and compatible with the double terminator. | + | Our new vector <partinfo>pSB1A7</partinfo> eliminates the read-through problems we had with <partinfo>pSB1A3</partinfo>, but is incompatible with double terminator <partinfo>B0015</partinfo>. We hope to design and build a new vector that is well-insulated from read-through and compatible with the double terminator. |
[[Image:dc_vector.jpg]] | [[Image:dc_vector.jpg]] | ||
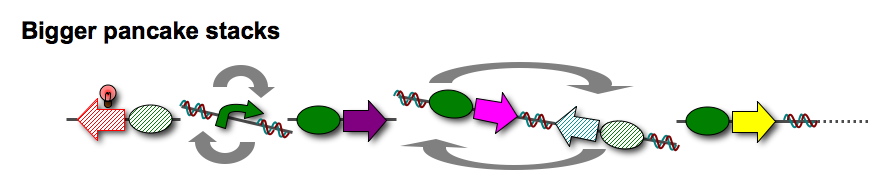
Finally, we want to build bigger stacks of pancakes, using additional antibiotic resistance genes and other functional units of DNA. | Finally, we want to build bigger stacks of pancakes, using additional antibiotic resistance genes and other functional units of DNA. | ||
| - | [[Image | + | [[Image:dc_stacks.jpg]] |
Latest revision as of 15:59, 1 December 2006
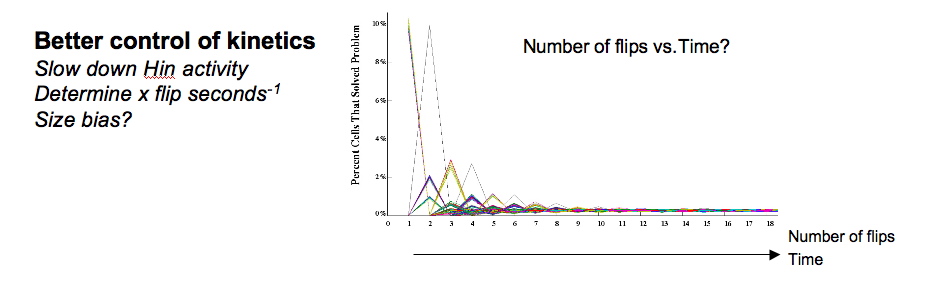
We have succesfully designed, modeled, and built a simple computer for solving the pancake problem. However, we do not yet have control over the production of Hin. Flipping occurs so quickly that we cannot calibrate our observations with our mathematical model. Once we can calibrate with the model, we will be able to determine flip rates and investigate whether shorter or longer segments of DNA are more likely to be flipped.
Our new vector eliminates the read-through problems we had with , but is incompatible with double terminator . We hope to design and build a new vector that is well-insulated from read-through and compatible with the double terminator.
Finally, we want to build bigger stacks of pancakes, using additional antibiotic resistance genes and other functional units of DNA.