PSB1A7 Cloning Solution
From 2006.igem.org
(→Solution to pSB1A7 cloning problem: Pancake stacks without B0015) |
(→Solution to pSB1A7 cloning problem: Pancake stacks without B0015) |
||
| (3 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
[[Image:Pancakes_noTT.gif|600px]] | [[Image:Pancakes_noTT.gif|600px]] | ||
| - | As shown above, we redesigned our two-pancake stack to exclude the double terminator at the end. So far, two permutations of this newly designed stack [(1,2) and (1,-2)]have been successfully cloned into pSB1A7. We've also eliminated the RE to "slown down" Hin invertase function. Construct #2 carries AraC (pBad repressor) and Hin on a second plasmid marked by kanamycin resistance. The | + | As shown above, we redesigned our two-pancake stack to exclude the double terminator at the end. So far, two permutations of this newly designed stack [(1,2) and (1,-2)]have been successfully cloned into pSB1A7. We've also eliminated the RE to "slown down" Hin invertase function. Construct #2 carries AraC (pBad repressor) and Hin on a second plasmid marked by kanamycin resistance. The double terminator upstream of Hin should block read-through transcription. PC-AraC (reverse) should be constitutively expressed and does not require insulation in our system. Plasmids #1 and #2 will be co-transformed into E. coli, and cells carrying both will be selected for using Amp + Kan media. |
Tests are currently under way to determine if the AraC/ Hin generator on plasmid #2 can | Tests are currently under way to determine if the AraC/ Hin generator on plasmid #2 can | ||
| - | <br>1. '''Suppress pBad-driven transcription''' - | + | <br>1. '''Suppress pBad-driven transcription''' - The AraC protein suppresses the pBad promoter. Adding arabinose (repressor of AraC) should induce transcription from pBad and confer tetracycline resistance when pBad is in the proper orientation relative to RBS-Tet. |
| - | <br>2. ''' | + | <br>2. '''Stimulate DNA inversion''' - Hin expression (induced by the addition of IPTG) should stimulate DNA flipping. This will be assayed by tet resistance in the presence of arabinose and by NheI restriction digests. |
| + | |||
| + | |||
| + | The [[Biological Equivalence Problem|next section]] describes how we address biological equivalence of permutations (1,2) and (-2,-1). | ||
Latest revision as of 19:11, 31 October 2006
Solution to pSB1A7 cloning problem: Pancake stacks without B0015
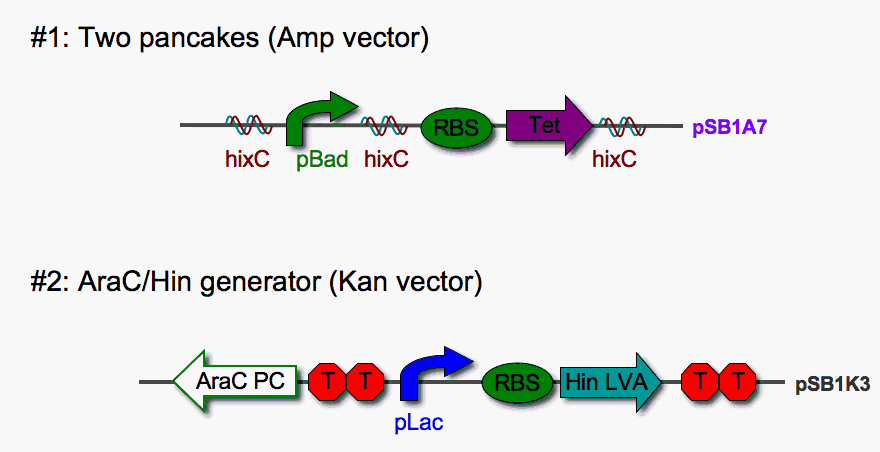
Our new cloning vector pSB1A7 insulates our parts from read-through transcription, but this vector does not accept parts that carry the B0015 double terminator. Our flipping device was originally designed with a B0015 near the end. Several attempts to clone parts carrying B0015 into pSB1A7 failed. This problem prompted a redesign of our system.
As shown above, we redesigned our two-pancake stack to exclude the double terminator at the end. So far, two permutations of this newly designed stack [(1,2) and (1,-2)]have been successfully cloned into pSB1A7. We've also eliminated the RE to "slown down" Hin invertase function. Construct #2 carries AraC (pBad repressor) and Hin on a second plasmid marked by kanamycin resistance. The double terminator upstream of Hin should block read-through transcription. PC-AraC (reverse) should be constitutively expressed and does not require insulation in our system. Plasmids #1 and #2 will be co-transformed into E. coli, and cells carrying both will be selected for using Amp + Kan media.
Tests are currently under way to determine if the AraC/ Hin generator on plasmid #2 can
1. Suppress pBad-driven transcription - The AraC protein suppresses the pBad promoter. Adding arabinose (repressor of AraC) should induce transcription from pBad and confer tetracycline resistance when pBad is in the proper orientation relative to RBS-Tet.
2. Stimulate DNA inversion - Hin expression (induced by the addition of IPTG) should stimulate DNA flipping. This will be assayed by tet resistance in the presence of arabinose and by NheI restriction digests.
The next section describes how we address biological equivalence of permutations (1,2) and (-2,-1).