PLac Tet Pancake Plan
From 2006.igem.org
m |
|||
| (4 intermediate revisions not shown) | |||
| Line 12: | Line 12: | ||
|- valign="top" | |- valign="top" | ||
| [[image:pLac_pancake1.gif|450px]] | | [[image:pLac_pancake1.gif|450px]] | ||
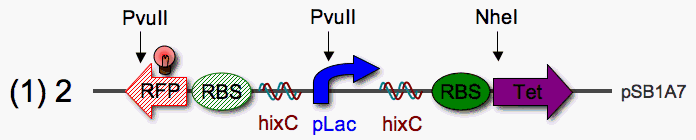
| - | | <b>Restriction Mapping</b>: PvuII sites occur once in the stationary RFP reporter and once in pLac. A PvuII digest can be used to check the orientation of pLac anywhere in the stack. Since PvuII and NheI share optimal buffers (Promega), a simple double digest could be used to check the orientation of | + | | <b>Restriction Mapping</b>: PvuII sites occur once in the stationary RFP reporter and once in pLac. A PvuII digest can be used to check the orientation of pLac anywhere in the stack. Since PvuII and NheI share optimal buffers (Promega), a simple double digest could be used to check the orientation of pLac and Tet relative to eachother.<br><br><b>Construction, Step 1</b>: First, I will build a hix-flanked pLac promoter (pLac single pancake) with a reverse RBS-RFP to the left and RBS-TetF to the right. If pLac can read through hix, the cells will be tetracycline resistant.<br><br> |
|- valign="top" | |- valign="top" | ||
| [[image:pLac_pancake2.gif|450px]] | | [[image:pLac_pancake2.gif|450px]] | ||
| Line 18: | Line 18: | ||
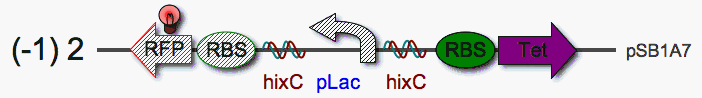
*indicate that Hin can invert a strong promoter (pLac). | *indicate that Hin can invert a strong promoter (pLac). | ||
*produce a reverse pLac (after flipping, plasmids will be purified and transformed into cells to produce clonal plasmids containing reverse pLac) | *produce a reverse pLac (after flipping, plasmids will be purified and transformed into cells to produce clonal plasmids containing reverse pLac) | ||
| - | *determine | + | *determine whether reverse pLac can promote reverse RBS-RFP expression |
| + | |- valign="top" | ||
| + | | [[image:pLac_Tet_pancakes.gif|450px]] | ||
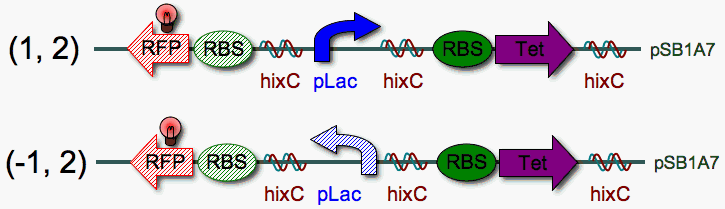
| + | | <b>Construction, Step 3</b>: A hix site will be added to the end of the unflipped and flipped pLac constructs to produce two-pancake stacks (1,2) and (-1,2). Using the same series of steps (1 through 3) with a reverse RBS-Tet, two additional stacks [(1,-2) and (-1,-2)] could be created.<br><br>In order to create the permutations with RBS-Tet in front of pLac [i.e. (2,1)], the reverse RBS-RFP reporter would have to be omitted so that more parts could be added to the left of RBS-Tet. | ||
|} | |} | ||
Latest revision as of 00:37, 27 February 2007
pBad Is a Weak Promoter
In our previous design, we aniticpated that a reverse RBS-RFP reporter would distinguish the biologically equivalent (1, 2) and (-2, -1) configurations of a pBad, TetR two-pancake stack. To test this, I placed the reverse RBS-RFP reporter to the left of (-2, -1). I also placed it to the left of (-1, -2) to see if the distance between the pBad promoter and the RFP would affect expression.
Only RFPrev-RBSrev-(-1, -2) confers weak expression of RFP (faint pink cell pellet), even after 0.2 - 2.0% arabinose induction in the absence of an extragenic copy of AraC (repressor of pBad). This result suggests that JM109 cells have endogenous AraC that represses pBad and that pBad is a weak promoter that requires close proximity to its coding region. Since one of our goals is to build long multi-coding sequence pancake stacks, pBad is a poor choice for this device (but may be useful in controlling Hin expression).
pLac Tet Pancake Assembly Plan
We've observed that pLac promotes strong expression of RBS-RFP, even in the absence of an inducer (IPTG). pLac may be better suited for our pancake stack device.