User:Irina Petrova
From 2006.igem.org
| Line 2: | Line 2: | ||
---- | ---- | ||
| - | I am a PhD student of the GRK 1305/1 “Plant Signal Systems” program in The Freiburg University[http://www.plant-signals.uni-freiburg.de/]. I am working on the detection and visualization of Arabidopsis thaliana root mRNA in Prof. Palme’s research group[http://www.biologie.uni-freiburg.de/forschung/botanik.php]. I am interested in bringing science and design together. I like DNA and iGEM Competition. | + | I am a PhD student of the GRK 1305/1 “Plant Signal Systems” program in The Freiburg University[http://www.plant-signals.uni-freiburg.de/]. I am working on the detection and visualization of Arabidopsis thaliana root mRNA in Prof. Palme’s research group[http://www.biologie.uni-freiburg.de/forschung/botanik.php]. I am interested in bringing science and design together. I like DNA and the iGEM Competition. |
---- | ---- | ||
| Line 15: | Line 15: | ||
The dress design is more interesting than a chip design (in my opinion ;). It is very individual and very fashionable. We want to follow fashion, don’t we? | The dress design is more interesting than a chip design (in my opinion ;). It is very individual and very fashionable. We want to follow fashion, don’t we? | ||
| - | On the | + | On the other hand, a broad range of variable forms can be important for an artificial life. I play with DNA like with my Barbie doll. |
The idea was to knit a nice blouse for Barbie without any boundary conditions. I used two methods of knitting: <br> | The idea was to knit a nice blouse for Barbie without any boundary conditions. I used two methods of knitting: <br> | ||
1) a rectilinear merge pattern, and <br> | 1) a rectilinear merge pattern, and <br> | ||
2) a staggered merge pattern <br> | 2) a staggered merge pattern <br> | ||
| - | + | following the terms of Paul Rothemund. The first one is simpler to understand; the second one is more practical for patterning. Only when you use a staggered merge pattern can you put all hairpins onto one side of the knitted DNA sheet with maximal density. | |
Have a look at the pictures: <br> | Have a look at the pictures: <br> | ||
| Line 32: | Line 32: | ||
[http://partsregistry.org/Part:BBa_J35001 BBa_J35001] | [http://partsregistry.org/Part:BBa_J35001 BBa_J35001] | ||
to create the Nike-logo pattern. <br> | to create the Nike-logo pattern. <br> | ||
| - | + | The other ones would be: | |
[http://partsregistry.org/Part:BBa_J35003 BBa_J35003], | [http://partsregistry.org/Part:BBa_J35003 BBa_J35003], | ||
[http://partsregistry.org/Part:BBa_J35004 BBa_J35004], | [http://partsregistry.org/Part:BBa_J35004 BBa_J35004], | ||
| Line 38: | Line 38: | ||
[http://partsregistry.org/Part:BBa_J35006 BBa_J35006], | [http://partsregistry.org/Part:BBa_J35006 BBa_J35006], | ||
[http://partsregistry.org/Part:BBa_J35007 BBa_J35007]. <br> | [http://partsregistry.org/Part:BBa_J35007 BBa_J35007]. <br> | ||
| - | Your | + | Your choice! |
---- | ---- | ||
Revision as of 14:50, 30 October 2006
I am a PhD student of the GRK 1305/1 “Plant Signal Systems” program in The Freiburg University[http://www.plant-signals.uni-freiburg.de/]. I am working on the detection and visualization of Arabidopsis thaliana root mRNA in Prof. Palme’s research group[http://www.biologie.uni-freiburg.de/forschung/botanik.php]. I am interested in bringing science and design together. I like DNA and the iGEM Competition.
email: irina.petrova(at)biologie.uni-freiburg.de
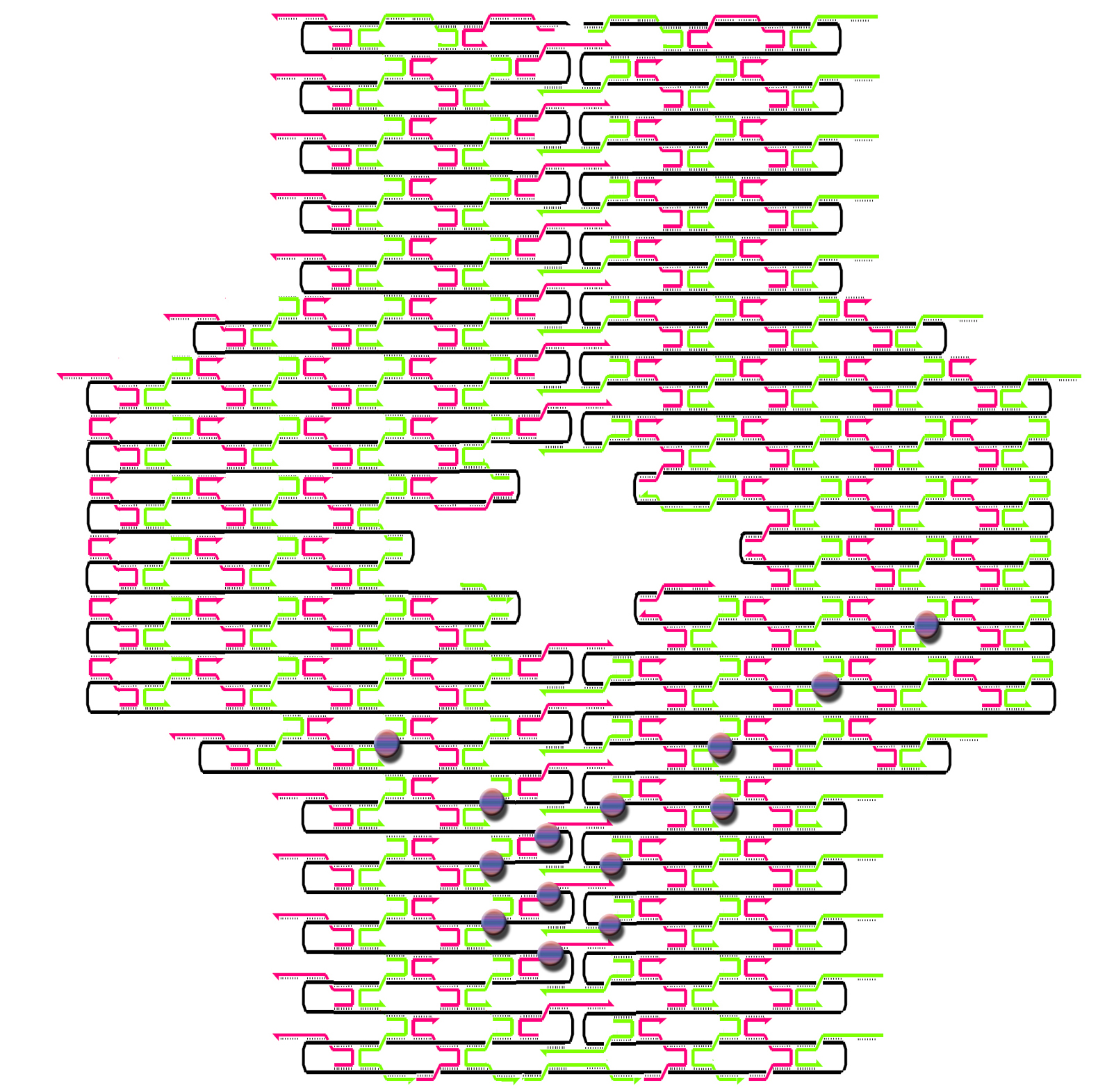
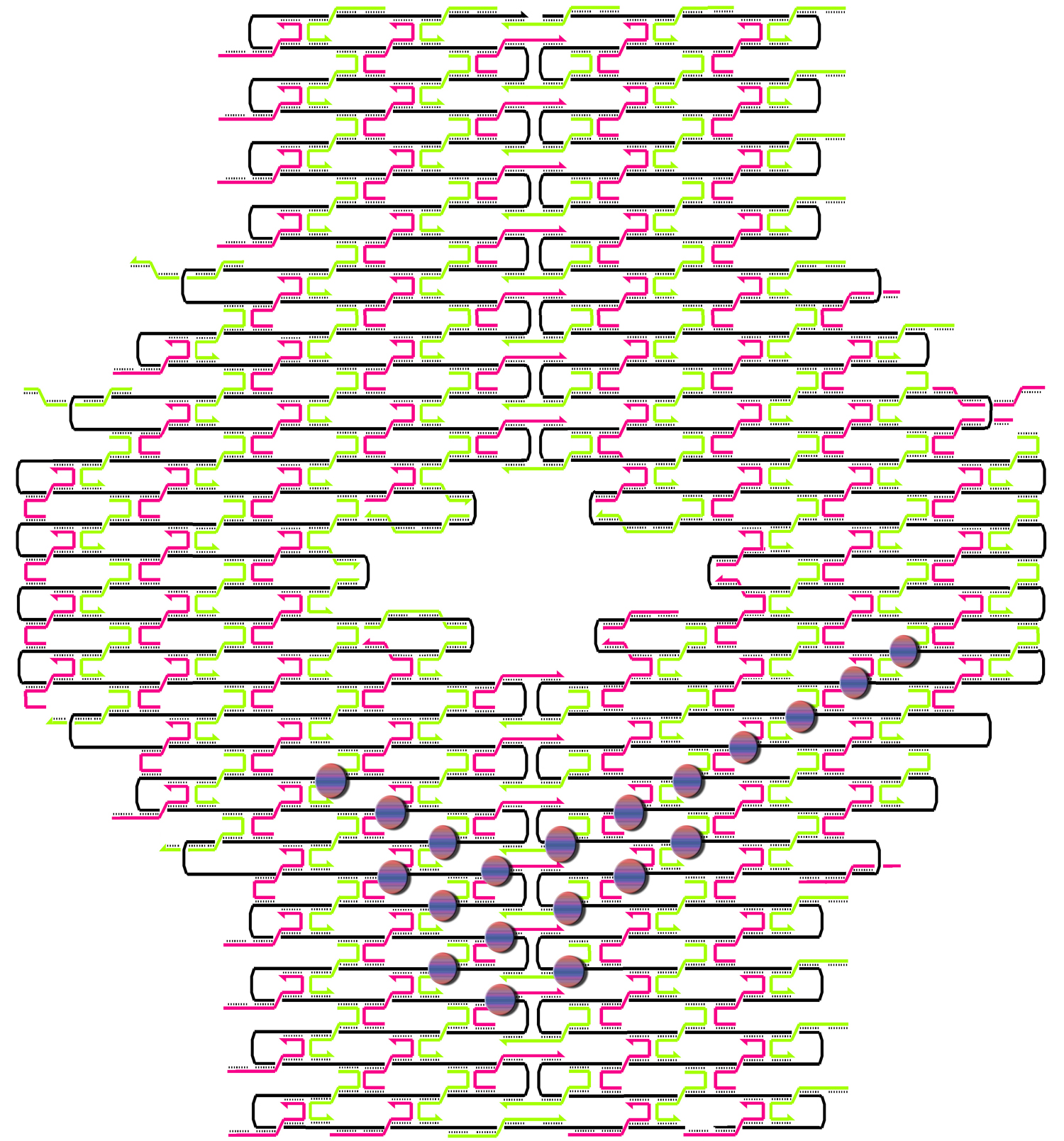
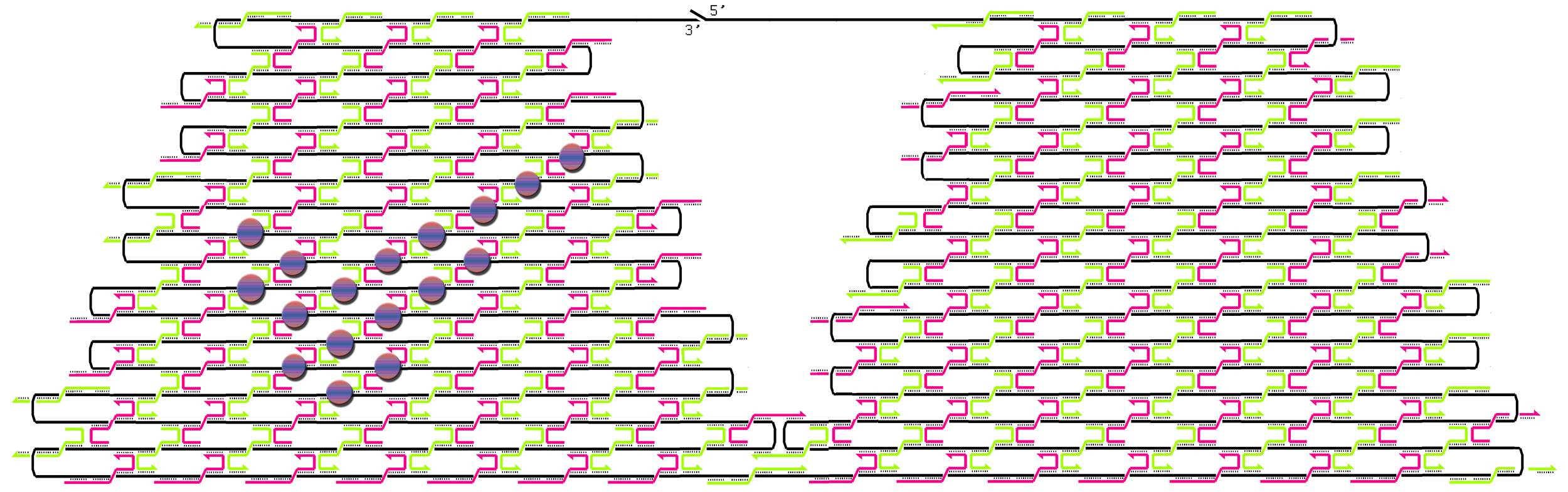
Individual project: Nike nano collection (Blouse and Skirt)
The dress design is more interesting than a chip design (in my opinion ;). It is very individual and very fashionable. We want to follow fashion, don’t we?
On the other hand, a broad range of variable forms can be important for an artificial life. I play with DNA like with my Barbie doll.
The idea was to knit a nice blouse for Barbie without any boundary conditions. I used two methods of knitting:
1) a rectilinear merge pattern, and
2) a staggered merge pattern
following the terms of Paul Rothemund. The first one is simpler to understand; the second one is more practical for patterning. Only when you use a staggered merge pattern can you put all hairpins onto one side of the knitted DNA sheet with maximal density.
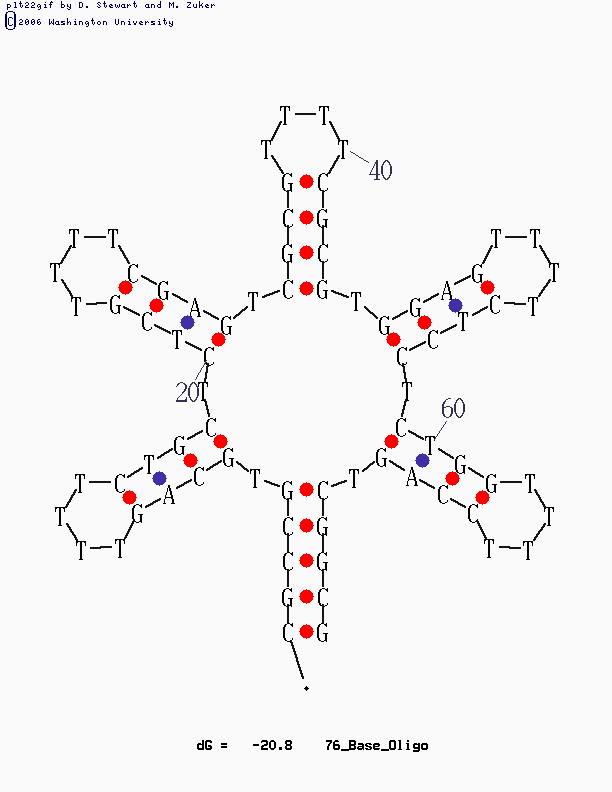
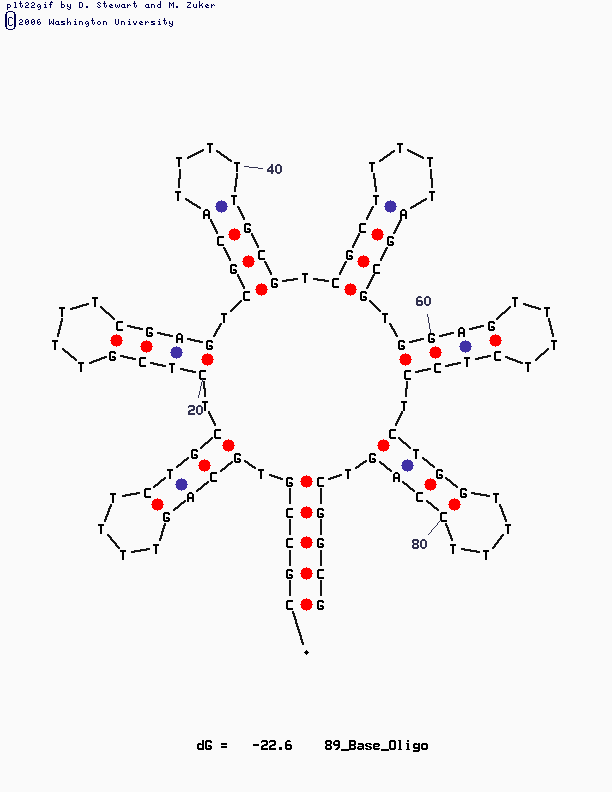
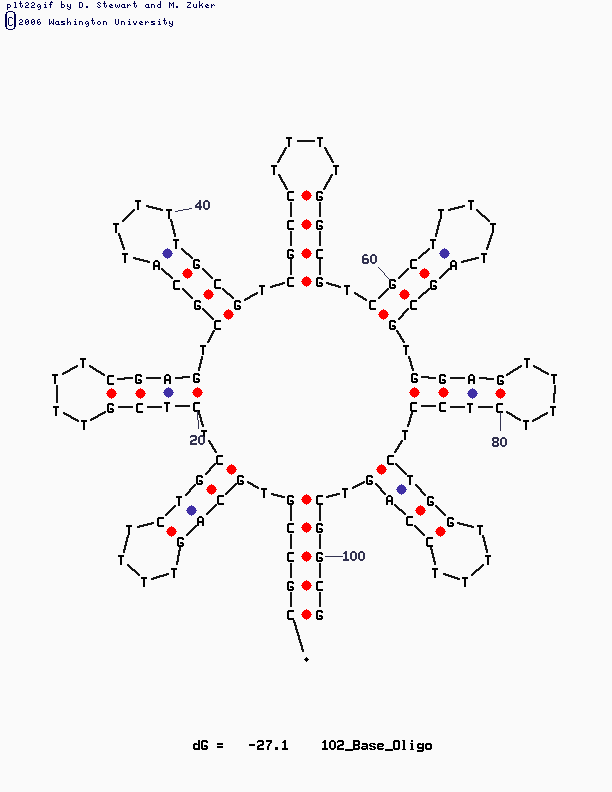
Have a look at the pictures:
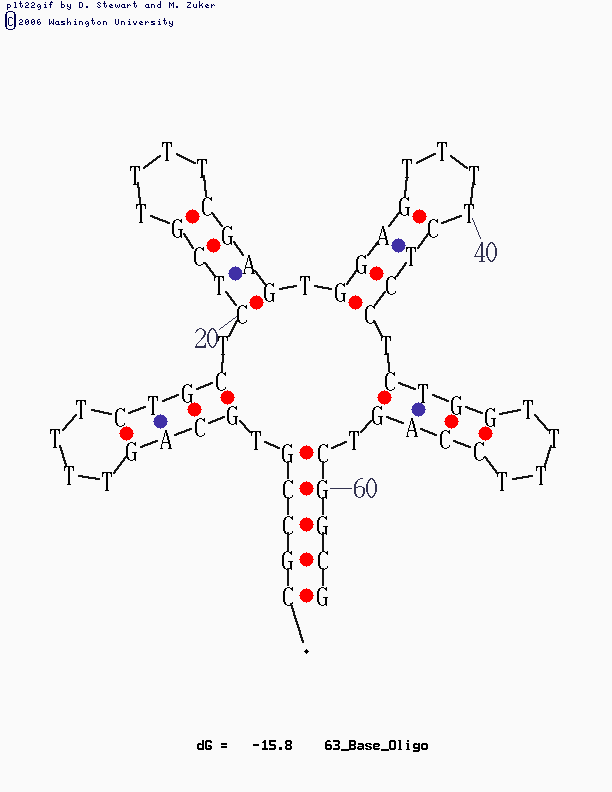
This design is for [http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/sequences/m13mp18.txt M13mp18] scaffold DNA. I use the fork hairpin
[http://partsregistry.org/Part:BBa_J35001 BBa_J35001]
to create the Nike-logo pattern.
The other ones would be:
[http://partsregistry.org/Part:BBa_J35003 BBa_J35003],
[http://partsregistry.org/Part:BBa_J35004 BBa_J35004],
[http://partsregistry.org/Part:BBa_J35005 BBa_J35005],
[http://partsregistry.org/Part:BBa_J35006 BBa_J35006],
[http://partsregistry.org/Part:BBa_J35007 BBa_J35007].
Your choice!
My photos will help you.
Another pretty possibility is the hybrids (color)FP with DNA-binding proteins that bind to specific staples, e.g. [http://partsregistry.org/Part:BBa_J35030 BBa_J35030]
[http://2006.igem.org/wiki/index.php/Freiburg_University_2006 Home]