Lab Work
From 2006.igem.org
| Line 1: | Line 1: | ||
| + | ===10th August 2006=== | ||
| + | Minipreps were prepared today from yesterday's transformations, and they will be done tomorrow. | ||
| + | |||
| + | ===9th August 2006=== | ||
| + | The ligation was done today, and retransformations of old stock DNA was started. These were plated. | ||
| + | |||
| + | In addition, experiments with the prepared arsenic constructs have revealed potential problems with the arsenic promoter sequence which will have to be checked. In addition, the '3U' lacZ gene apparently does not work at all, so the 4U one will be used from now on. | ||
| + | |||
| + | ===8th August 2006=== | ||
| + | The 5G plasmid was cut with EcoRI and XbaI,and the plasmid containing the terminator with EcoRI and SpeI in preparation for ligation. They were run on a gel and extracted, and then frozen for tomorrow. | ||
| + | |||
===More stuff to be done=== | ===More stuff to be done=== | ||
| Line 104: | Line 115: | ||
| - | === | + | ===7th August 2006=== |
Of the 12 minipreps prepared for the ars + 4U(lacZ) constructs, only one appeared to have been sucessful, and this was preserved by plating and overnight incubation till tomorrow. Tomorrow the ars + 3U and ars + 4U constructs will be compared to verify. | Of the 12 minipreps prepared for the ars + 4U(lacZ) constructs, only one appeared to have been sucessful, and this was preserved by plating and overnight incubation till tomorrow. Tomorrow the ars + 3U and ars + 4U constructs will be compared to verify. | ||
Revision as of 13:16, 10 August 2006
10th August 2006
Minipreps were prepared today from yesterday's transformations, and they will be done tomorrow.
9th August 2006
The ligation was done today, and retransformations of old stock DNA was started. These were plated.
In addition, experiments with the prepared arsenic constructs have revealed potential problems with the arsenic promoter sequence which will have to be checked. In addition, the '3U' lacZ gene apparently does not work at all, so the 4U one will be used from now on.
8th August 2006
The 5G plasmid was cut with EcoRI and XbaI,and the plasmid containing the terminator with EcoRI and SpeI in preparation for ligation. They were run on a gel and extracted, and then frozen for tomorrow.
More stuff to be done
- Urease
Once we've got the urease clone that Chris is working on, we need to see if it still works in the strain of bacteria that we're using. So we need to transfer the plasmid into our strain, then do some tests on it to see if it works.
If it does work, we need to mutate out the inappropriate sites. The protocol for doing this is online at http://www.stratagene.com/manuals/200518.pdf The primers for it will be ordered once we know that the urease works, and they are as follows:
For mutating out EcoRI:
cttcggcaaagccgagttcagcttcatatccag
ctggatatgaagctgaactcggctttgccgaag
For mutating out SpeI:
cctctgatttccactagagagaccgttttctgctc
gagcagaaaacggtctctctagtggaaatcagagg
Once we've done that, the urease is ready to be biobricked, and also
ready to be assembled with our shiny new promoter (once the promoter is
ready).
- LacI/Lambda cI promoter:
The primers for this have been designed and should be ordered today.
They are as follows.
For the cI binding site:
Primer 1: 5'- catctcgagattatcaccgccagagg -3' (has Xho1 site added
on 5' end)
Primer 2: 5'- gtggagctccgttaaatctatcaccgc - 3' (has Sac1 site
added on 5' end)
These should be used for a PCR reaction with the Lambda bacteriophage
that Chris found.
For the lacI/lacZ:
We should have a primer left over with the suffix added from the time
we were biobricking LacZ. The other primer is
5' gttctcgagaattgtgagcggataac 3' (with XhoI site added on 5' end)
These should be used for a PCR reaction with an E. coli cell.
Provided the PCR works, we'll end up with the following two chunks of
DNA:
---SacI site---lambda cI binding site+promoter---XhoI site---
---XhoI site-LacI binding site---LacZ---SpeI site---
There is a new Biobrick vector Chris designed (named Edinbrick I :) ),
with a SacI site added after the prefix, which is the vector we'll be
using for this reaction.
Cut the vector and the two fragments with SacI, XhoI and SpeI, ligate
them together, and hopefully *fingers crossed* we'll have our shiny
new biobricked promoter.
- Testing the promoter:
Because the cI binding site is being transferred in its entirety, we
will assume that the cI will bind to it and repress it properly. So all
we need to check is:
-That the promoter works. By putting it into an E. coli strain that's
missing lacI and lacZ, to see if the lacZ attached to the promoter is
expressed. This can be done quickly if we put x-gal into the growth
medium - leave the cells to grow and see if they turn blue.
-That the promoter is repressed by LacI. Put the construct into an E.
coli cell that expresses LacI constitutively, but is missing the lacZ
gene. Then grow one set of colonies on a medium that contains IPTG +
x-gal (should allow lacZ expression) and another on a medium containing
x-gal, but no IPTG (shouldn't allow lacZ expression). If all goes
according to plan, the colonies on the first one should be blue, and on
the second one should be white.
7th August 2006
Of the 12 minipreps prepared for the ars + 4U(lacZ) constructs, only one appeared to have been sucessful, and this was preserved by plating and overnight incubation till tomorrow. Tomorrow the ars + 3U and ars + 4U constructs will be compared to verify.
The lambda cI (5G) plasmid from the iGEM plates was successfully transformed into E.coli, but there were problems with the lamda cI promoter (9C) plasmid: few colonies grew upon transformation, and upon analysis on the gel the bands indicating the plasmid were very faint.
To be done in the following days
To do list for while Judith is away - she expects everything to be finished on her return from Serbia ;)
Primers ordered for lambda cI promoter, and the urease. These can be transformed into cells, once they have been PCR-ed.
The lambda cI promoter needs to be biobricked beside the urease gene.
ArsR which we have already biobricked with lacZ can be biobricked beside the lambda cI repressor protein to result in a construct which has an alkaline pH when arsenic is not present and a neutral/acidic pH when arsenic is present.
Upstairs, tests tests tests.
So.
- PCR using primers for lambda cI promoter and urease gene.
- Digests and minipreps etc. to obtain two constructs, one with the arsenic promoter and the lambda cI repressor and one with the urease gene and the lambda cI promoter.
- Both constructs need to be in the same cell. Could put them all in a row or on two different plasmids.
4th August 2006
Ligations of the Ars promoter insert with the 4U lacZ plasmid were repeated today in the hopes of getting it to work this time around. The ligated plasmid was then transformed into competent cells and plated, and individual cultures will be prepared during the weekend.
In addition, two new plasmids from the iGEM plates were transformed and plated :
| 9C | BBa_R0051 | Repressor, lambda cI | Plate 1 | AmpR |
| 5G | BBa_C0051 | Promoter, lambda cI regulated | Plate 1 | AmpR |
On Monday it will be seen if any of these transformations have worked, and hopefully our lucky streak will come back.
3rd August 2006
Minipreps were carried out on the new colonies containing the 4U plasmid with the Ars insert, and the gel showed that the ligation did not work in any of these colonies either. We are digesting another 4U vector with EcoRI and XbaI to see if we can obtain a working construct for this form of the lacZ gene.
We do however, have two plates of colonies of the Ars insert sucessfully ligated by the 3U version of the lacZ. Get in. Now to see if they work.
2nd August 2006
Minipreps were carried out on last night's cultures, and the inserts cut out with EcoRI and PstI. Gel analysis revealed that the ligations were not very successful (there goes our lucky streak in the lab), but 2 of the minipreps were successful and those colonies were streaked on to fresh agar plates. Some more colonies (12) will be grown overnight for further miniprepping tomorrow.
Tentatively, we can say we have our simple construct for the arsenic biosensor project ready to test.
1st August 2006
12 colonies were picked from yesterday's transformations and placed into media for overnight incubation.
31st July 2006
The ars insert and the lacZ and terminator containing vector were isolated from a gel, and ligated then transformed into E. coli. A test gel of the lacZ fragment from the original vector, and the lacZ fragment hopefully containing the terminator showed the first ligation was successful. Finally, today's ligation was transformed into E.coli.
28th July 2006
6 minipreps were carried out for each of the two transformations done yesterday, and the inserts were cut out with EcoRI and PstI, the gel showed the ligations were successful, so preparative digestions were carried out on the ars fragment with EcoRI and SbaI and on each of the lacZ fragments with EcoRI and XbaI.
27th July 2006
A large number of colonies were produced from yesterday's work, so 12 cultures were set up from each of the two transformations to be incubated overnight, with minipreps to be done tomorrow.
26th July 2006
Digestions were carried out on the 1I terminator containing vector with EcoRI and XbaI, and on the LacZ inserts with EcoRI and SpaI. These fragments were ran on a gel, and isolated from the gel to ligate and transform colonies with the LacZ plus terminator plasmid.
21st July 2006
8 minipreps each were carried out on the ars and the 2 lacZ plasmids (24 total), along with 2 for vector with no insert as controls. The inserts were then cut out of the plasmids with the EcoRI and the PstI restriction endonucleases. These were run on a gel to identify successful cut fragments, and these will be used next week.
20th July 2006
The transformed colonies grew sucessfully. 8 individual colonies of each were placed in LB+ampicillin media for growing overnight.
19th July 2006
Ars and LacZ PCR fragments with sticky ends were cleaned up, and the vector isolated from a gel. Ligations were set up for two of the lacZ inserts, and the ars insert with a control of no insert. These were transformed into E.coli and plated.
18th July 2006
Hot weather resulted in general laziness and angst amongst iGEM team. Apart from that, the vector which contained the terminator, and the PCR fragments for Ars genes and new lacZ gene were digested with XbaI and PstI to be ligated tomorrow.
17th July 2006
24 minipreps were carried out on individual colonies that grew after being transformed with the (suspected) ligations between 7K and 30 and 9E and 1I, and cut with EcoRI and PstI to find if the ligation was successful on a gel. The 7K and 30 ligations didn't appear to have been successful, but the 9E and 1I ligation looked to have worked.
14th July 2006
The PCR products from yesterday were ran on a gel, and strong bands found at around 600bp for the arsenic parts, as expected, and these were extracted from the gel. The lacZ products resulted in two distinct bands for each different strain, so all four were isolated and extracted from the gel.
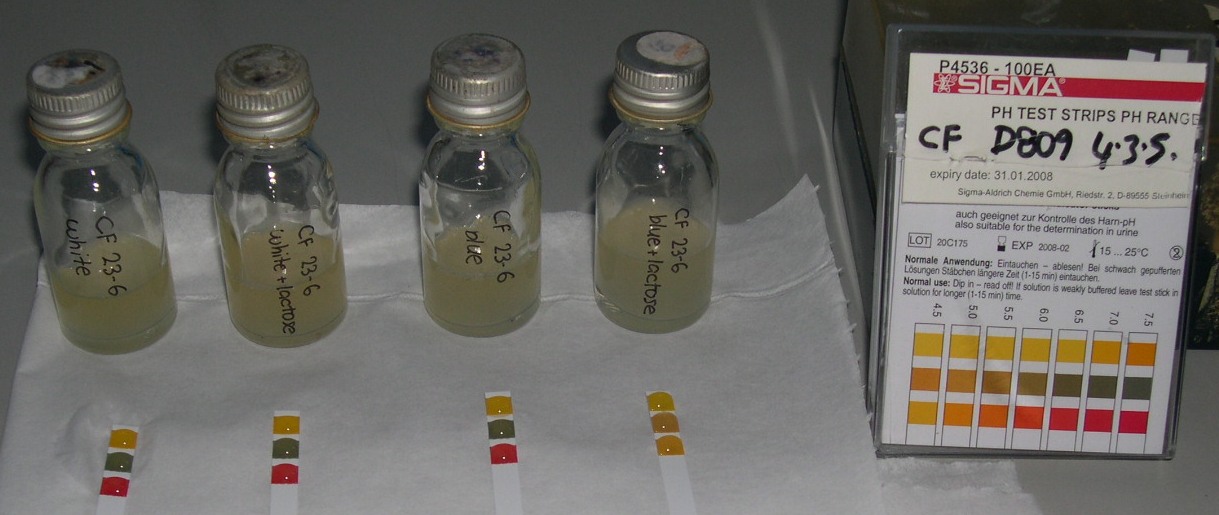
Conclusions from pH lab work
- Experiments 1 & 2 indicate that we are using the wrong growth medium, if we want to achieve a neutral pH without lactose present
- A response is indicated after 3.5 - 4 hours, but 90% of final state is achieved somewhere between 400 - 600 min
- In presence of LB medium, without lactose, the pH stabilises at about 8.5
- The maximum pH change was registered at 4.5, from 8.5 to 4.0
- just adding water to the cultures instead of growth medium slows the response time
- Experiment 3 indicates that a 50% culture to growth medium ratio increases the response time
- Experiment 4 indicates that there is a threshold level of lactose necessary for the acid transformation to occur. Once all the lactose has been used, the bacteria revert to converting amino acids to ammonia, raising the pH
- Other than the threshold level of lactose, the concentration of lactose and culture has no effect on the speed or strength of the response.
13th July 2006
We performed ligations with the DNA purified from the colonies transformed on the 6th, 7K (promoter) to 30 (RBS) and 9E (lacZ) to 1I (terminator). The recombinant plasmids were transformed (hopefully) into competent cells and plated on medium containing Xgal and IPTG, with a pBluescript colony also plated as a control. We also suspended more original colonies in liquid culture as a backup.
The primers ordered for a better lacZ gene, and the arsR and ars promoter arrived and we did PCR with cells of two different E. coli strains.
In order to determine the ideal conditions to achieve a pH response, three different growth mediums were established:
| Parameter: | 50% culture | 25% culture | 12.5% culture |
| Ampicillin: | 50 μl | 50 μl | 50 μl |
| IPTG: | 50 μl | 50 μl | 50 μl |
| Culture: | 25 ml | 12.5 ml | 6.25 ml |
| Sterile water: | 25 ml | 37.5 ml | 43.75 ml |
From these cultures we achieved the following results:
| Parameter: | 50% culture (pH) | 25% culture (pH) | 12.5% culture (pH) |
| Time (in min) | |||
| 0 | 8.48 | 8.47 | 8.43 |
| 30 | 8.28 | 8.17 | 7.97 |
| 60 | 8.18 | 8.05 | 7.85 |
| 90 | 7.99 | 7.83 | 7.62 |
| 120 | 7.67 | 7.61 | 7.61 |
| 150 | 7.53 | 7.49 | 7.54 |
| 180 | 7.48 | 7.47 | 7.47 |
| 210 | 7.38 | 7.34 | 7.38 |
| 240 | 7.33 | 7.33 | 7.46 |
At the end of the experiment the pH meter was tested using pH 7.0 and pH 4.0 buffers. The 4.0 buffer was measured at 3.42 and the 7.0 buffer measured 7.21. Although there is a degree of inaccuracy in the measurements, the electrode was still functioning correctly after the experiments.
12th July 2006
The pH response over time was again measured but in this experiment, we used liquid cultures which were already saturated (i.e. in the stationary phase).
| Parameter: | Blue saturated culture | White saturated culture |
| Ampicillin: | 50 μl | 50 μl |
| IPTG: | 50 μl | 50 μl |
| Culture: | 50 ml | 50 ml |
The results were as follows:
| Parameter: | Blue culture (pH) | White culture (pH) |
| Time (in min) | ||
| 0 | 8.51 | 8.41 |
| 15 | 8.41 | 8.31 |
| 30 | 8.33 | 8.24 |
| 45 | 8.32 | 8.25 |
| 60 | 8.26 | 8.28 |
| 90 | 8.05 | 8.28 |
| 120 | 7.79 | 8.34 |
| 150 | 7.57 | 8.36 |
| 180 | 7.37 | 8.33 |
| 210 | 7.11 | 8.45 |
| 270 | 6.60 | 8.49 |
| 1350 | 4.03 | 8.30 |
11th July 2006
Today we tested the timed pH response in 2 cell cultures: 1 with the LacZ gene, which was activated by IPTG, and one that had this gene absent.
We cut the isolated plasmid DNA with restriction enzymes to remove the inserts from the promoter and lacZ part, and open the vectors for the RBS and terminator. We ran gels, and these showed that the restriction had succeeded, but had not yielded much DNA. The correct bands were cut out of the gel to purify the inserts and vectors, ready for ligation.
10th July 2006
The colonies transformed on the 6th made it this time, and we isolated the plasmid DNA from three individual colonies for each biobrick. We also transformed some E. coli with
| 23E | pSB1A3 | Plasmid | Plate 1 | AmpR |
to serve as an empty plasmid for the new LacZ and arsenic promoter/repressor parts which we will create.
7th July 2006
When bacteria with the lacZ gene inserted are present in a medium containing lactose, the pH does drop significantly.
6th July 2006
Unfortunately the colonies we plated on the 4th did not survive due to a problem with the competent cells we used, so today we repeated transforming and plating colonies containing the following parts:
| 9E | BBa_E0033 | LacZ alpha | Plate 2 | KanR |
| 1I | BBa_B0015 | Terminator | Plate 1 | AmpR |
| 7K | BBa_R0010 | IPTG responsive promoter | Plate 1 | AmpR |
| 3O | BBa_B0034 | RBS | Plate 1 | AmpR |
4th July 2006
We plated colonies containing plasmids with the following parts:
| 9E | BBa_E0033 | LacZ alpha | Plate 2 | KanR |
| 3P | BBa_0010 | Terminator | Plate 2 | AmpR |
| 7K | BBa_R0010 | IPTG responsive promoter | Plate 1 | AmpR |
[http://2006.igem.org/Standard_Protocols Standard Protocols]
[http://2006.igem.org/University_of_Edinburgh_2006 Main page]