Berkeley2006-PromoterMain
From 2006.igem.org
JCAnderson (Talk | contribs) |
JCAnderson (Talk | contribs) |
||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
| - | |||
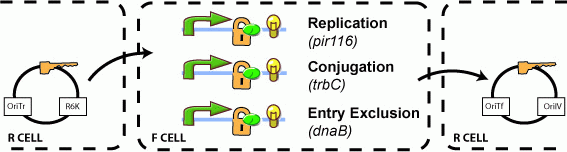
'''The ability to maintain, send, and receive plasmids is lock-controlled''' | '''The ability to maintain, send, and receive plasmids is lock-controlled''' | ||
| + | '''For addressable conjugation within a network, ''' | ||
| + | |||
| + | |||
[[Image:Berkeley2006_Conjugation7.GIF]] | [[Image:Berkeley2006_Conjugation7.GIF]] | ||
| + | === === | ||
| + | It is almost always the case that when combining a PoPS signal with an actuation device, such as bacterial conjugation, the rates of transcription and translation must be tuned to match the levels necessary for activation of the device. In other words, too much protein in the "off" state can lead to a situation where the observed phenotype occurs independent of the key. Similarly, too little protein in the "on" state can result in a scenario where the bacteria do not conjugate even when the key is present. When controlling a gene transcriptionally, this matching of rates can be achieved by manipulating the ribosome binding site of the actuation device. In translation control, however, the sequence of the ribosome binding site is an essential feature of the mode of regulation and cannot be changed independently. In this scenario, the transcription rate of the device must instead be changed. | ||
| + | |||
| + | '''Isolatation of a family of constitutive promoter parts from a combinatorial library to vary transcription rates''' | ||
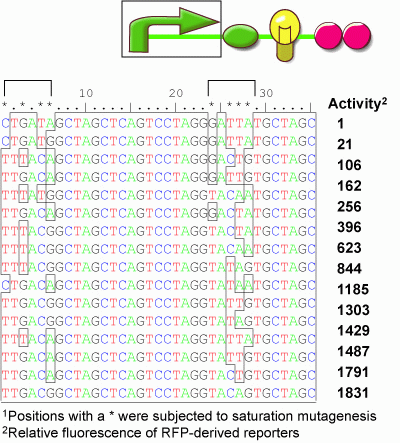
We thereore developed a set of transcriptional promoter parts that vary the transcription rate. The matching of rates can then be achieved by identifying a promoter providing the proper transcription rate to match the systems requirements. To make these parts, we adopted a combinatorial approach. A synthetic promoter sequence was targetted with saturation mutagenesis employing a binary swap at 8 specific positions within the -10 and -35 sequences. In this way, 8 positions were simultaneously varied to be either the consensus sequence for that position, or not. We constructed the library fused to an RFP reporter gene containing the strong ribosome binding site b0034, the mRFP 1 gene (E0###), and a double terminator (b0015). We inserted the 256 member library into the RFP reporter plasmid and picked 96 visibly white colonies and 96 visibly red colonies. The cells were grown to saturation and then assayed by cytometry. From this screen, we identified 18 unique promoter variants that each display a different level of fluorescense activity spanning the physiological range between the level observed with cells containing no mRFP gene and values higher than those observed with a Ptet promoter (r0040). These promoter parts will find general use in tuning the expression level of Biobrick devices. | We thereore developed a set of transcriptional promoter parts that vary the transcription rate. The matching of rates can then be achieved by identifying a promoter providing the proper transcription rate to match the systems requirements. To make these parts, we adopted a combinatorial approach. A synthetic promoter sequence was targetted with saturation mutagenesis employing a binary swap at 8 specific positions within the -10 and -35 sequences. In this way, 8 positions were simultaneously varied to be either the consensus sequence for that position, or not. We constructed the library fused to an RFP reporter gene containing the strong ribosome binding site b0034, the mRFP 1 gene (E0###), and a double terminator (b0015). We inserted the 256 member library into the RFP reporter plasmid and picked 96 visibly white colonies and 96 visibly red colonies. The cells were grown to saturation and then assayed by cytometry. From this screen, we identified 18 unique promoter variants that each display a different level of fluorescense activity spanning the physiological range between the level observed with cells containing no mRFP gene and values higher than those observed with a Ptet promoter (r0040). These promoter parts will find general use in tuning the expression level of Biobrick devices. | ||
Revision as of 21:07, 28 October 2006
The ability to maintain, send, and receive plasmids is lock-controlled For addressable conjugation within a network,
It is almost always the case that when combining a PoPS signal with an actuation device, such as bacterial conjugation, the rates of transcription and translation must be tuned to match the levels necessary for activation of the device. In other words, too much protein in the "off" state can lead to a situation where the observed phenotype occurs independent of the key. Similarly, too little protein in the "on" state can result in a scenario where the bacteria do not conjugate even when the key is present. When controlling a gene transcriptionally, this matching of rates can be achieved by manipulating the ribosome binding site of the actuation device. In translation control, however, the sequence of the ribosome binding site is an essential feature of the mode of regulation and cannot be changed independently. In this scenario, the transcription rate of the device must instead be changed.
Isolatation of a family of constitutive promoter parts from a combinatorial library to vary transcription rates
We thereore developed a set of transcriptional promoter parts that vary the transcription rate. The matching of rates can then be achieved by identifying a promoter providing the proper transcription rate to match the systems requirements. To make these parts, we adopted a combinatorial approach. A synthetic promoter sequence was targetted with saturation mutagenesis employing a binary swap at 8 specific positions within the -10 and -35 sequences. In this way, 8 positions were simultaneously varied to be either the consensus sequence for that position, or not. We constructed the library fused to an RFP reporter gene containing the strong ribosome binding site b0034, the mRFP 1 gene (E0###), and a double terminator (b0015). We inserted the 256 member library into the RFP reporter plasmid and picked 96 visibly white colonies and 96 visibly red colonies. The cells were grown to saturation and then assayed by cytometry. From this screen, we identified 18 unique promoter variants that each display a different level of fluorescense activity spanning the physiological range between the level observed with cells containing no mRFP gene and values higher than those observed with a Ptet promoter (r0040). These promoter parts will find general use in tuning the expression level of Biobrick devices.
Next Section: Logic Computation in Cellular Networks