Harvard 2006
From 2006.igem.org
ZhipengSun (Talk | contribs) (→The Harvard University 2006 iGEM Team) |
ShawnDouglas (Talk | contribs) |
||
| Line 1: | Line 1: | ||
| - | + | __NOTOC__ | |
| + | {| {{table}} | ||
| + | |- | ||
| + | | colspan="3" align="center" style="color:#fff; background:#cc0000;"|Harvard University 2006 iGEM Team | ||
| + | |- | ||
| + | | rowspan="2" | [[Image:HarvardTeamPhoto2006.jpg|left|300px]] | ||
| + | | valign="top" style="width:120px;" |'''Students''' | ||
| + | *Tiffany Chan | ||
| + | *Katherine Fifer | ||
| + | *Lewis Hahn | ||
| + | *Hetmann Hsieh | ||
| + | *Jeffrey Lau | ||
| + | *Valerie Lau | ||
| + | *Matthew Meisel | ||
| + | *David Ramos | ||
| + | *Zhipeng Sun | ||
| + | *Perry Tsai | ||
| + | | valign="top" style="width:180px;" | | ||
| + | '''Teaching Fellows''' | ||
| + | *[http://openwetware.org/wiki/User:Doucette Chris Doucette] | ||
| + | *[http://openwetware.org/wiki/User:ShawnDouglas Shawn Douglas] | ||
| + | *[http://openwetware.org/wiki/User:Stroustr Nicholas Stroustrup] | ||
| + | '''Faculty Advisors''' | ||
| + | *[http://arep.med.harvard.edu/ George Church] (HMS) | ||
| + | *[http://www.eecs.harvard.edu/~rad/ Radhika Nagpal] (DEAS) | ||
| + | *[http://hst.mit.edu/biosketch/Shah.html Jagesh Shah] (HMS) | ||
| + | *[http://research2.dfci.harvard.edu/shih/ William Shih] (HMS) | ||
| + | *[http://silver.med.harvard.edu/ Pamela Silver] (HMS) | ||
| + | |} | ||
| + | ==The Harvard University 2006 iGEM Team== | ||
| + | This year Harvard's team consisted of 10 undergraduate students, with backgrounds in molecular and cellular biology, biochemistry, and computer science. With the help of five faculty advisors and three graduate-level teaching advisors, they devised and executed three separate projects. | ||
| + | 1. Construction of novel DNA nanostructures for the purpose of stealth drug delivery, | ||
| + | 2. Exploration of cell-surface targeting using interchangable and linkable aptamers (adaptamers) and Lpp-OmpA fusion vehicle for bacterial surface display. | ||
| - | + | 3. Reconstitution of a circadian oscillator from cyanobacteria into E. coli, | |
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | ==DNA Nanobox: A Device for Stealth Drug Delivery== | ||
| + | *[http://openwetware.org/wiki/IGEM:Harvard/2006/DNA_nanostructures openwetware page] | ||
| - | + | '''Project Overview''' | |
| - | * | + | *Our goal is to design and implement molecular containers, which can be dynamically opened and closed by an external stimulus. |
| - | * | + | *The containers will be implemented as DNA nanostructures, which afford a significant degree of positional control and chemical versatility. |
| - | * | + | *As an initial proof-of-concept, we plan to use our DNA containers to demonstrate controllable activation ("delivery") of anti-thrombin aptamers. |
| - | + | *We expect that molecular containers could have several interesting scientific and clinical applications, such as | |
| + | **Drug and gene delivery | ||
| + | **Bio-marker scavenging (early detection of biomarkers) | ||
| + | **Directed evolution (compartmentalized selections) | ||
| + | **Using multiplexing for combinatorial chemical synthesis | ||
| + | **Capture and stabilization of multiprotein complexes | ||
| + | **Protein folding (chaperones) | ||
| + | **Cell sorting | ||
| - | == | + | '''Results''' |
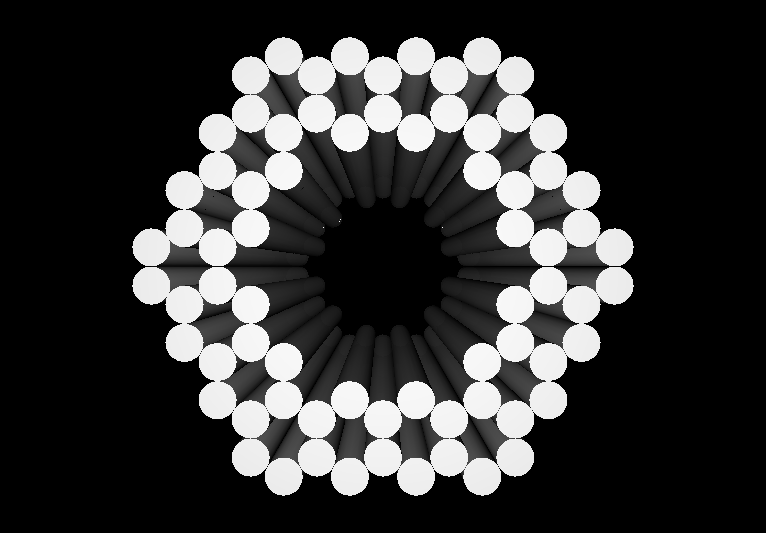
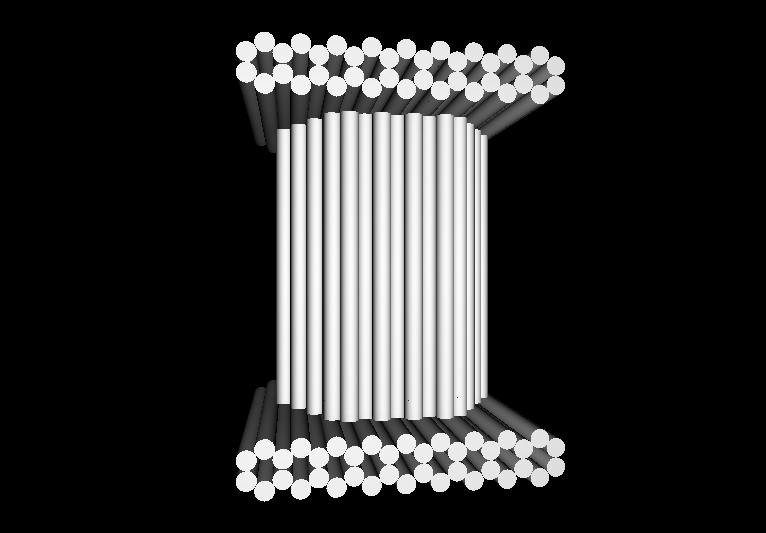
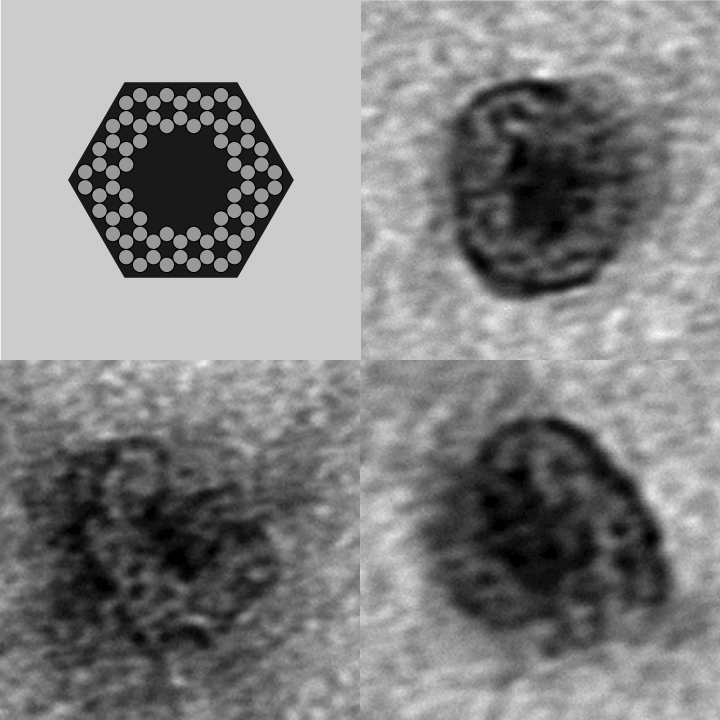
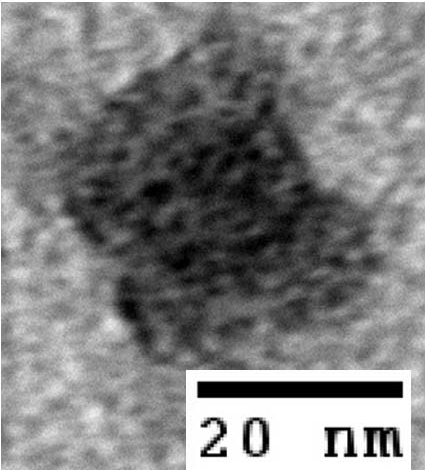
| - | *[http:// | + | <gallery> |
| - | *[http:// | + | Image:Harvard2006-nanobox-3Dview1.jpg|3D Cross Section |
| - | + | Image:Harvard2006-nanobox-3Dview2.jpg|3D cartoon of Design | |
| - | *[http:// | + | Image:Harvard2006-nanobox-EM1.jpg|Negative stain EM |
| - | *[http:// | + | Image:Harvard2006-nanobox-EM2.png|EM side view |
| + | </gallery> | ||
| + | |||
| + | |||
| + | '''Future Plans''' | ||
| + | |||
| + | ==Cell Surface Targeting== | ||
| + | *[http://openwetware.org/wiki/IGEM:Harvard/2006/Cell_surface_targeting openwetware page] | ||
| + | |||
| + | '''Adaptamers''' | ||
| + | *[http://openwetware.org/wiki/IGEM:Harvard/2006/Adaptamers openwetware page] | ||
| + | |||
| + | '''Project Overview''' | ||
| + | *We are interested in the mostly equivalent problems of targeting substrates to cells and cells to substrates. In the former case, targeting a substrate to the cell would facilitate uptake and a subsequent cellular response, important in fields such as drug delivery. Directing cells to particular places, for instance, a column, could be used to isolate cells and perform diagnostics. | ||
| + | *Briefly, we are pursuing two methods to attack this problem. In one route, we will express streptavidin on the E. coli cell surface, hence providing a target for any biotinylated molecule. In the second, we will build on the work of Tahiri-Alaoui et al. (2002) in developing bi-specific DNA "adaptamers" that can bind a cell surface and a substrate. | ||
| + | |||
| + | '''Results''' | ||
| + | |||
| + | '''Future Plans''' | ||
| + | |||
| + | '''Cell Surface Fusion Proteins''' | ||
| + | *[http://openwetware.org/wiki/IGEM:Harvard/2006/Fusion_proteins openwetware page] | ||
| + | |||
| + | '''Project Overview''' | ||
| + | |||
| + | '''Results''' | ||
| + | |||
| + | '''Future Plans''' | ||
| + | |||
| + | ==A Circadian Oscillator for E.coli== | ||
| + | *[http://openwetware.org/wiki/IGEM:Harvard/2006/Cyanobacteria openwetware page] | ||
| - | + | '''Project Overview''' | |
| - | + | ||
| - | + | '''Results''' | |
| - | + | '''Future Plans''' | |
| - | == | + | ==Complete Electronic Notebooks== |
| - | + | Detailed records of our summer activities, including results, can be found on openwetware, which the team used to host its [http://openwetware.org/wiki/IGEM:Harvard/2006 electronic notebook]. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 02:00, 29 October 2006
| Harvard University 2006 iGEM Team | ||
Students
|
Teaching Fellows
Faculty Advisors
| |
The Harvard University 2006 iGEM Team
This year Harvard's team consisted of 10 undergraduate students, with backgrounds in molecular and cellular biology, biochemistry, and computer science. With the help of five faculty advisors and three graduate-level teaching advisors, they devised and executed three separate projects.
1. Construction of novel DNA nanostructures for the purpose of stealth drug delivery,
2. Exploration of cell-surface targeting using interchangable and linkable aptamers (adaptamers) and Lpp-OmpA fusion vehicle for bacterial surface display.
3. Reconstitution of a circadian oscillator from cyanobacteria into E. coli,
DNA Nanobox: A Device for Stealth Drug Delivery
- [http://openwetware.org/wiki/IGEM:Harvard/2006/DNA_nanostructures openwetware page]
Project Overview
- Our goal is to design and implement molecular containers, which can be dynamically opened and closed by an external stimulus.
- The containers will be implemented as DNA nanostructures, which afford a significant degree of positional control and chemical versatility.
- As an initial proof-of-concept, we plan to use our DNA containers to demonstrate controllable activation ("delivery") of anti-thrombin aptamers.
- We expect that molecular containers could have several interesting scientific and clinical applications, such as
- Drug and gene delivery
- Bio-marker scavenging (early detection of biomarkers)
- Directed evolution (compartmentalized selections)
- Using multiplexing for combinatorial chemical synthesis
- Capture and stabilization of multiprotein complexes
- Protein folding (chaperones)
- Cell sorting
Results
Future Plans
Cell Surface Targeting
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Cell_surface_targeting openwetware page]
Adaptamers
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Adaptamers openwetware page]
Project Overview
- We are interested in the mostly equivalent problems of targeting substrates to cells and cells to substrates. In the former case, targeting a substrate to the cell would facilitate uptake and a subsequent cellular response, important in fields such as drug delivery. Directing cells to particular places, for instance, a column, could be used to isolate cells and perform diagnostics.
- Briefly, we are pursuing two methods to attack this problem. In one route, we will express streptavidin on the E. coli cell surface, hence providing a target for any biotinylated molecule. In the second, we will build on the work of Tahiri-Alaoui et al. (2002) in developing bi-specific DNA "adaptamers" that can bind a cell surface and a substrate.
Results
Future Plans
Cell Surface Fusion Proteins
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Fusion_proteins openwetware page]
Project Overview
Results
Future Plans
A Circadian Oscillator for E.coli
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Cyanobacteria openwetware page]
Project Overview
Results
Future Plans
Complete Electronic Notebooks
Detailed records of our summer activities, including results, can be found on openwetware, which the team used to host its [http://openwetware.org/wiki/IGEM:Harvard/2006 electronic notebook].