ETH2006 and
From 2006.igem.org
(→Implementation alternatives) |
(→Modeling) |
||
| Line 24: | Line 24: | ||
=== Modeling === | === Modeling === | ||
''AND-tRNA'' → [[ETH_Sim_Mod_AND_tRNA|simulation results]] / [[ETH_Sens_And|sensitivity analysis]] | ''AND-tRNA'' → [[ETH_Sim_Mod_AND_tRNA|simulation results]] / [[ETH_Sens_And|sensitivity analysis]] | ||
| + | |||
| + | ==== Current simulation results ==== | ||
| + | {| | ||
| + | |- | ||
| + | | align="center" | ''topside view, ht<sub>Prot</sub>=2h'' || align="center" | ''3d surface plot, ht<sub>Prot</sub>=2h'' || align="center" | ''3d surface plot, ht<sub>Prot</sub>=5h'' | ||
| + | |- | ||
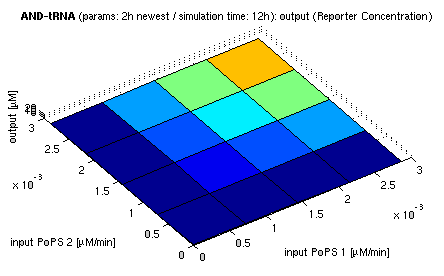
| + | | [[image:ETH_Sim_And_2h_flat.png|300px|center|topside view of simulation results for AND gate, protein halflife period: 2h]] | ||
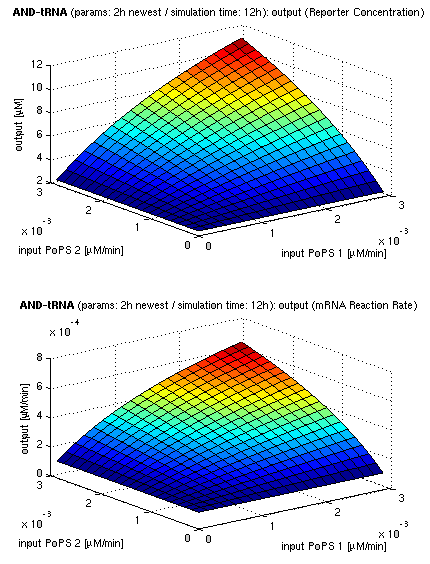
| + | | [[image:ETH_Sim_And_2h.png|350px|center|3d surface plot of simulation results for AND gate, protein halflife period: 2h]] | ||
| + | | [[image:ETH_Sim_And_5h.png|350px|center|3d surface plot of simulation results for AND gate, protein halflife period: 5h]] | ||
| + | |- | ||
| + | | colspan="3" align="center" | ''simulation results for AND gate, different protein halflife periods<br/>top / bottom: output (z axis) is mRNA rate (∼PoPS) / reporter concentration''<br/>(x/y: ''inputs'', z: ''output'', ht<sub>Prot</sub>: ''protein halflife period'') | ||
| + | |} | ||
| + | |||
| + | For every plot, 25 (left) / 100 (middle, right) ODE simulations were performed, simulating a time period of 12h. Simulations with 2h / 5h halflife period for proteins have been run, showing little difference in the outcome, though. Complete parameter allocation can be found in the [[ETH_Simulation_Scripts|matlab scripts]] as well as the system of ordinary differential equations (ODEs). | ||
| + | |||
| + | {{ETH Sim Input Rate}} | ||
| + | |||
| + | Amplifying/damping the input rates by small constant factors has influence on the qualitative outcome of the simulation. | ||
| + | * it is thus important to know how strong the input of the gate has to be. | ||
| + | * we can regulate this by choosing/designing the predecessor gate accordingly or | ||
| + | * by changing the ribosome binding sites to strengthen/weaken the input signal. | ||
| + | |||
| + | We accounted for this by adding restriction enzyme sites to the DNA. | ||
| + | |||
| + | In order to find out about other important, say ''sensitive'' parameters, we wanted to do this more systematically. The keyword has already been given: sensitivity analisys. | ||
=== Assembly procedure === | === Assembly procedure === | ||
Revision as of 16:11, 29 October 2006
Contents |
AND gate
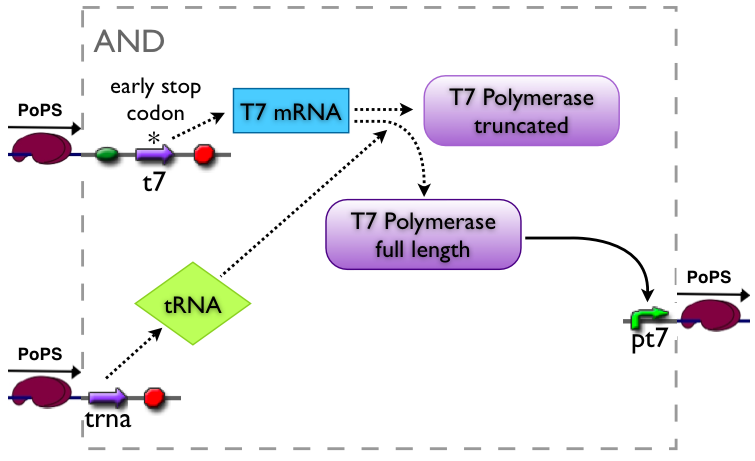
The AND gate's PoPs output activity should be correlated to the PoPs input activity as shown in the picture:
input A ^
| L D H
| D D D
| L D L
+--------->
input B
output: High, Low, Dont care
Implementation alternatives
Part to be used in the prototype system: [http://partsregistry.org/Part:BBa_J34100 BBa_J34100]
An AND gate produces an output only if both input signals are present. We want to implement such a “device” by using a suppressor tRNA. In our proposed model Signal 1 would lead to the transcription of the T7 RNA polymerase. However, an early stop codon was introduced into the coding sequence, leading to the expression of a truncated, non-functional protein. Signal 2, on the other hand, will lead to the expression of a suppressor tRNA. This tRNA recognizes the stop codon, preventing the end of translation and leading to incorporation of a glutamine into the nascent amino acid chain, so that translation can be completed. Consequently, a functional T7 RNA polymerase is present which will recognize its cognate promoter (T7 promoter) and therefore initiate expression of the downstream reporter gene (GFP).
Modeling
AND-tRNA → simulation results / sensitivity analysis
Current simulation results
| topside view, htProt=2h | 3d surface plot, htProt=2h | 3d surface plot, htProt=5h |
| simulation results for AND gate, different protein halflife periods top / bottom: output (z axis) is mRNA rate (∼PoPS) / reporter concentration (x/y: inputs, z: output, htProt: protein halflife period) | ||
For every plot, 25 (left) / 100 (middle, right) ODE simulations were performed, simulating a time period of 12h. Simulations with 2h / 5h halflife period for proteins have been run, showing little difference in the outcome, though. Complete parameter allocation can be found in the matlab scripts as well as the system of ordinary differential equations (ODEs).
The input rates (PoPS) have been chosen in the range of mRNA transcription rate, which was estimated according to the following assumptions:
- E.coli cytoplasm volume is approximately 6.7*10-16 l
- average number of mRNA molecules: 10
→ concentrationmRNA = 10/(6.7*10-16 * 6.022*1023) M = 0.0248 μM - at equilibrum, mRNA rate and degredation balance each other. Assuming half life period of 30min for mRNA, the result is
→ ratemRNA = concentrationmRNA * log(2)/30 μM/min = 5.7265e-04 μM/min
Amplifying/damping the input rates by small constant factors has influence on the qualitative outcome of the simulation.
- it is thus important to know how strong the input of the gate has to be.
- we can regulate this by choosing/designing the predecessor gate accordingly or
- by changing the ribosome binding sites to strengthen/weaken the input signal.
We accounted for this by adding restriction enzyme sites to the DNA, so that we can cut out current RBS to replace it by another of different strength.
Amplifying/damping the input rates by small constant factors has influence on the qualitative outcome of the simulation.
- it is thus important to know how strong the input of the gate has to be.
- we can regulate this by choosing/designing the predecessor gate accordingly or
- by changing the ribosome binding sites to strengthen/weaken the input signal.
We accounted for this by adding restriction enzyme sites to the DNA.
In order to find out about other important, say sensitive parameters, we wanted to do this more systematically. The keyword has already been given: sensitivity analisys.
Assembly procedure
Test procedure
Test results
Parts
[http://partsregistry.org/Part:BBa_J34100 BBa_J34100]