University of Texas 2006
From 2006.igem.org
| Line 55: | Line 55: | ||
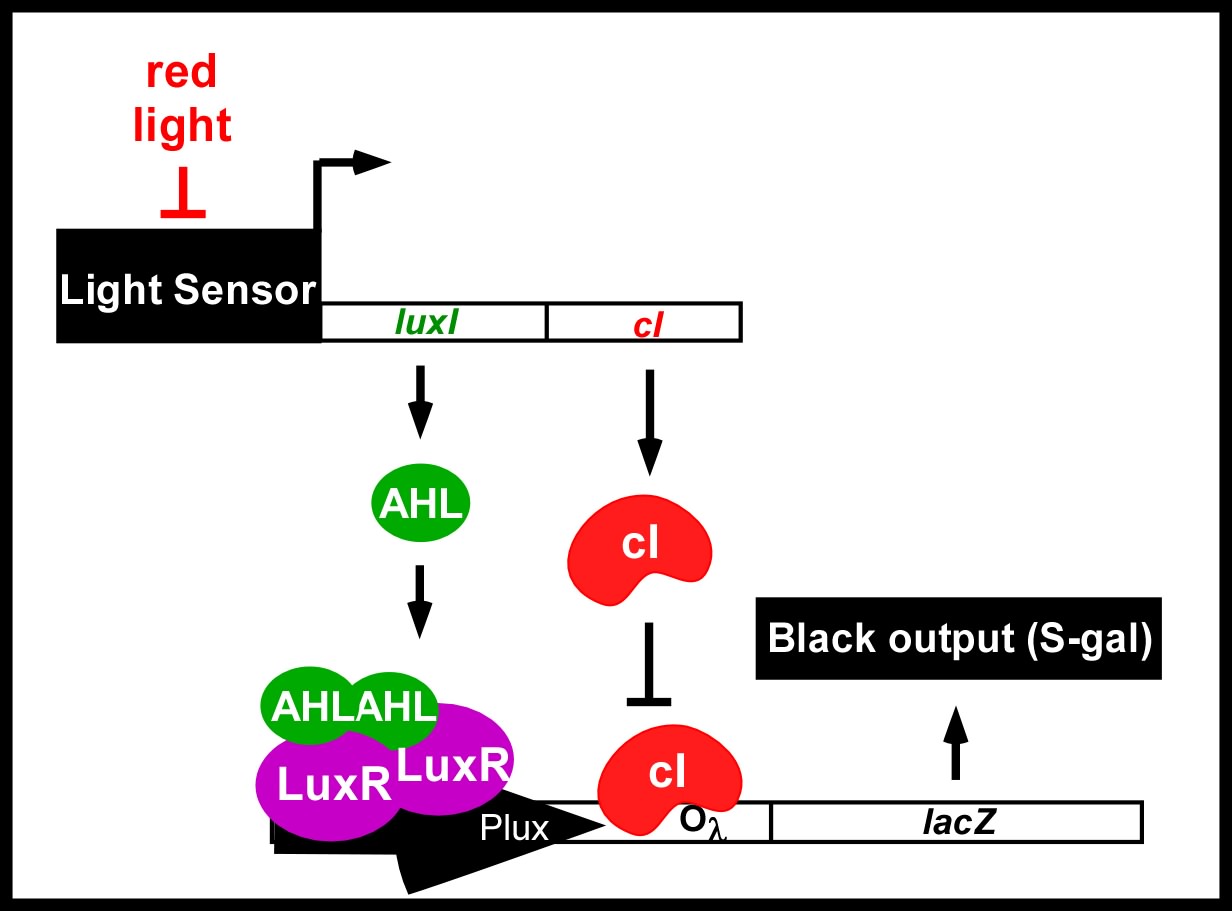
<div>This was the first step in engineering the edge detector. We have now connected the light-responsive circuitry to the remainder of the edge detection circuit ([http://partsregistry.org/Part:BBa_I15022 I15022]) the schematic and operation of which are shown in the figures below.</div> | <div>This was the first step in engineering the edge detector. We have now connected the light-responsive circuitry to the remainder of the edge detection circuit ([http://partsregistry.org/Part:BBa_I15022 I15022]) the schematic and operation of which are shown in the figures below.</div> | ||
| - | <div>[[Image:jt_1.jpg|thumb|left|400px| Edge Detector Circuit. | + | <div>[[Image:jt_1.jpg|thumb|left|400px| Edge Detector Circuit. The light sensing machinery from above has been black-boxed and the edge detection circuitry has been added downstream. Red light represses the expression of 2 genes; a biosynthetic gene for a membrane diffusible quorum sensing activator (AHL), and a dominant transcriptional repressor (cI).]]</div> |
<div> | <div> | ||
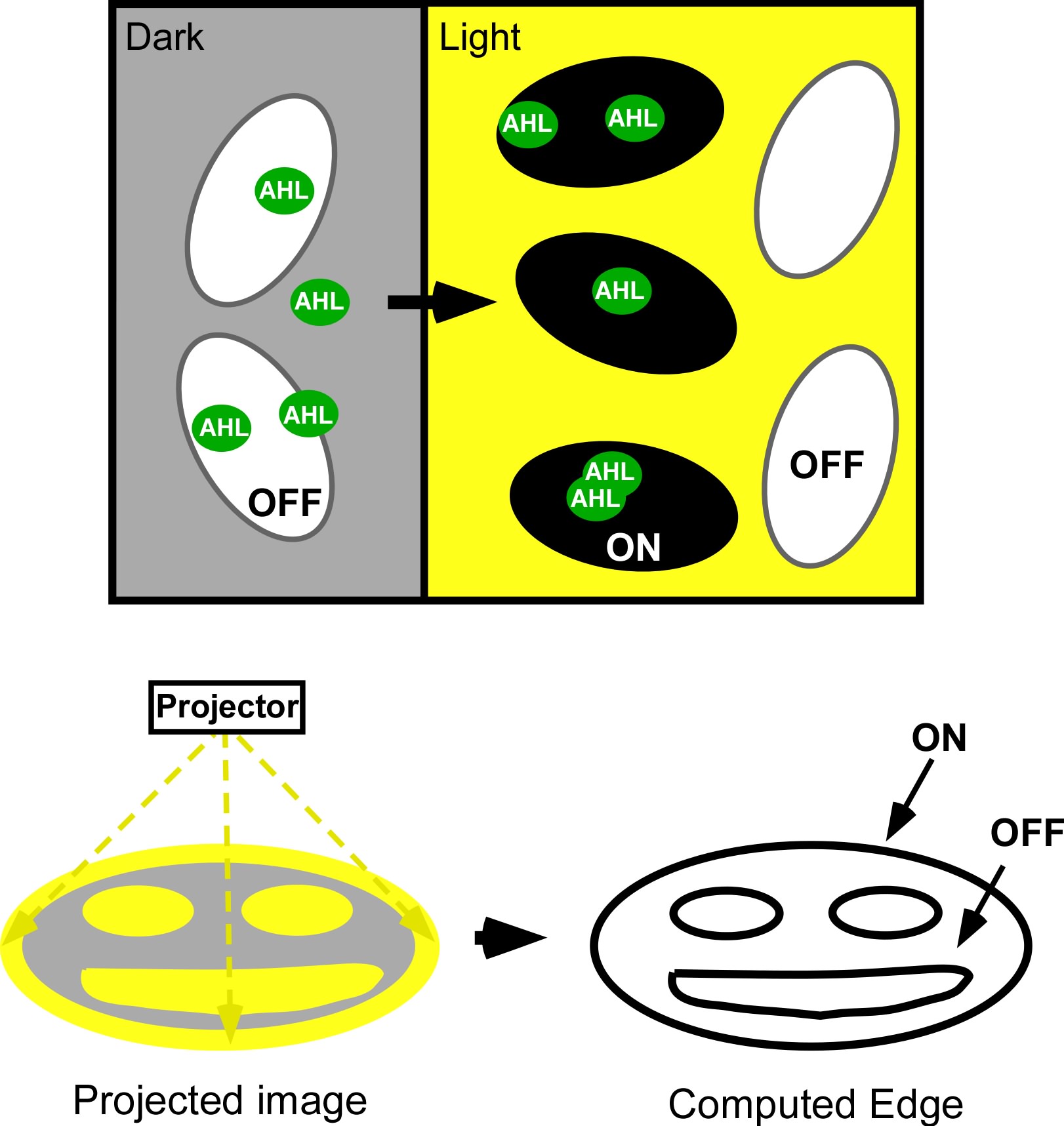
| - | [[Image:jt_2.jpg|thumb|right|300px| | + | [[Image:jt_2.jpg|thumb|right|300px| (Top) Micro-scale edge detection. (Bottom) Macro-scale edge detection. An image is projected onto an agar plate of ''E.coli'' expressing the circuitry shown to the left. Cells residing in dark areas express AHL but are dominantly repressed by CI. Illuminated cells express neither AHL nor cI. If they neighbor a dark area, they will be activated by locally diffusing AHL. In this way, only cells at the light dark boundary express a reporter, and the edge is detected.]] |
</div> | </div> | ||
Revision as of 19:08, 1 November 2006
The UT Austin team is:
- Aaron Chevalier
- Eric Davidson
- [http://openwetware.org/wiki/User:Jeff_Tabor Jeff Tabor]
- Laura Lavery
- Matt Levy
- [http://www.mine-control.com/ Zack Booth Simpson]
- Bryan Kaehr
Advisors:
- [http://ellingtonlab.org/ Andy Ellington]
- [http://polaris.icmb.utexas.edu/home.html Edward Marcotte]
Contents |
Previous work: Bacterial Photography
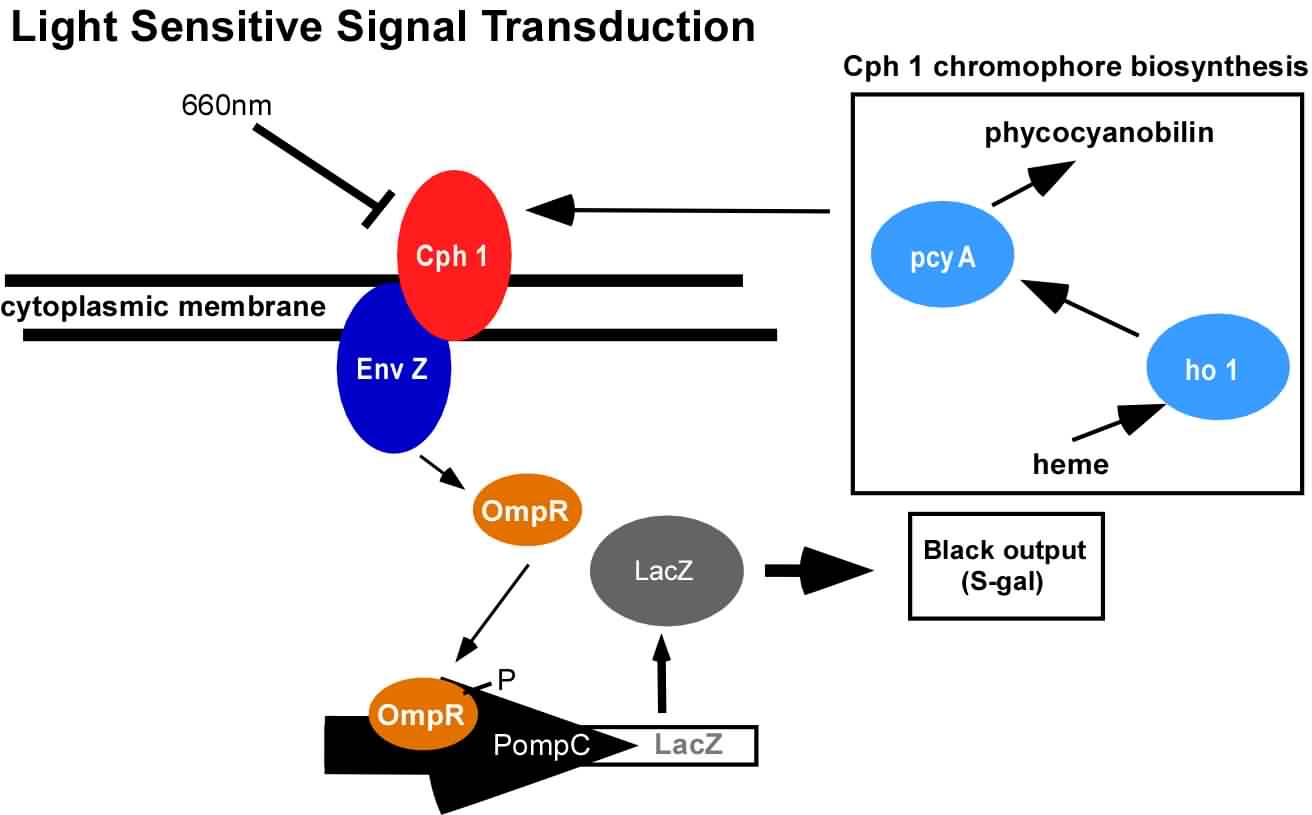
An obvious hurdle in the implementation of this system was genetically encoding light detection in the naturally blind E.coli. To accomplish this, we used a synthetic part engineered in the Voigt lab. This part, Cph8, ([http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=5302 I15010]) is an engineered fusion between a cyanobacterial light sensing phytochrome (Cph1) and an E.coli transmembrane histidine kinase, (EnvZ). 660nm light causes an isomerization in the Cph1 domain of the chimera which inactivates the histidine kinase acitity of EnvZ. When EnvZ is inhibited, a phosphorelay cascade which activates transcription from the OmpC promoter [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=3910 R0082]) and inhibits transcription from the OmpF promoter ([http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=3915 R0084]). We then demonstrated that when this system is expressed in E.coli, it is possible to transform each cell on an agar surface into a decision making pixel capable of reporting whether it is in the light or dark. The community of cells is therefore capable of genetically reproducing a light image.
Current work: Edge detector
Favorite Parts
- [http://partsregistry.org/Part:BBa_I15008 I15008]
- [http://partsregistry.org/Part:BBa_I15009 I15009]
- [http://partsregistry.org/Part:BBa_I15010 I15010]
- [http://partsregistry.org/Part:BBa_I15022 I15022]