Synthetic Counter (iGem2005 ETH Zurich)
From 2006.igem.org
Abstract. We report here the design and implementation in vivo of a gene circuit that can count up to 4. In essence, it uses two toggle switches, each storing 1 bit, to keep track of the 4 states. The design of the counter is highly modular, with the hope that it can be included as a unit in larger circuits, and also combined with further counter instances to keep track of a much larger number of states, up to (2^(n+1)) with n units. To facilitate further developments and integration to other projects, the counter is available in form of BioBricks. Among many exciting applications, the availability of a counter enables the execution of sequential instructions, and paves the way for the execution of artifical programs inside living cells.
Contents |
Introduction
The past few years have seen the emergence of the field of synthetic biology, in which functional units are designed and built into cells to generate a particular behaviour, and ultimately to better understand Life's mechanisms. Previous efforts include the creation of gene circuits that generate oscillating behaviour (Elowitz00), toggle switch functionality (Atkinson03), artificial cell-cell communication (Bulter04) or pattern-forming behaviour (Basu2005). The present document describes the design and realization of a gene circuit that counts to 4.
Design of the Counter
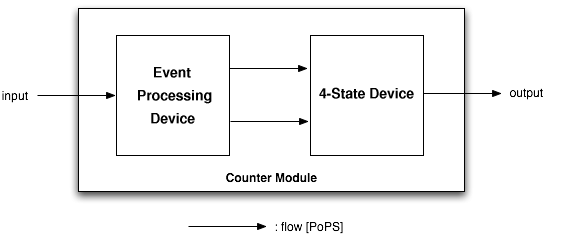
The counter is a genetic circuit that has 1 input and 4 outputs. It uses the input signal to switch from one of the four output to the next. When the input signal is high, either output 1 or 3 is active, when it is low, output 2 or 4 is active. Thus, output 1 and 3 alternatively keep track of high input signal, while output 2 and 4 alternatively keep track of low input signals.
As depicted above, the counter is made of two parts, serially linked:
- the "Input" module, which splits the input into two opposite signals.
- the "NOR" module, which uses these two signals to sequencially switch through the outputs 1, 2, 3 and 4.
Note that all interfaces have flows described in Polymerase Per Second (PoPS), is explained in details on the [http://partsregistry.org/cgi/htdocs/AbstractionHierarchy/index.cgi abstraction hierarchy] of the MIT Registry of Parts.
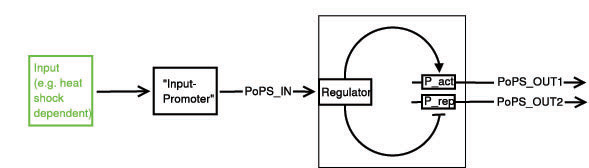
Input Module
The input module has 2 system boundaries, both of which are characterized by PoPS (Polymerase Per Second). One of the outputs should be high and the other low when S is high and vice versa when S is low.
PoPS_outn
^
¦
high ¦ PoPS_out2 _______ _________ PoPS_out1
¦ \ /
¦ \ /
¦ \ /
¦ \ /
¦ \ /
¦ X
¦ / \
¦ / \
¦ / \
¦ / \
low ¦ PoPS_out1 _______/ \_________ PoPS_out2
¦--------------------------------------------------> PoPS_in, t
0 1
Schematic
Below a preliminary parts-view of the module, i.e. encapsulation of biological specific implementations into a functional box with general PoPS interfaces.
With this design, any input can be used, as long as a promoter can be found that is either activated or suppressed by it. At the output, any kind of genes can be added which will be produced depending on the PoPS of that particular input.