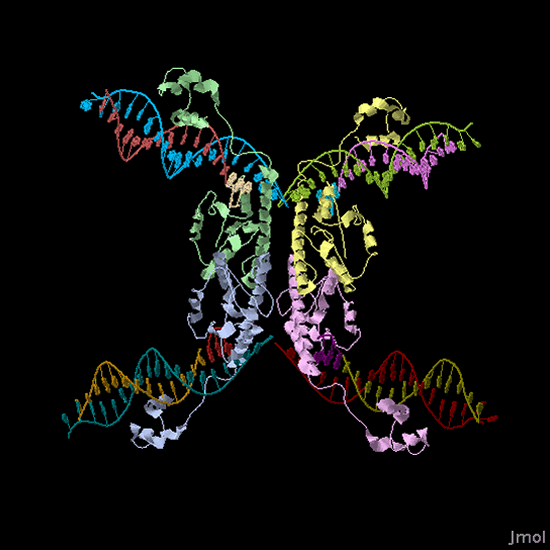Simulation results for two pancakes, useful for calibrating kinetics of pancake flipping. Our approach is to
model the problem by representing a pancake stack mathematically and biologically. A stack of pancakes can be represented by a series of integers in a certain order (ie. 1, 2, 3 vs. 3, 2, 1) where orientation is represented by a positive or negative value (1 vs. -1). Similarly, a series of DNA segments has a certain order (ie. promoter, coding region 1, coding region 2) and each unit has two possible orientations (plus strand vs. minus strand).
Approach
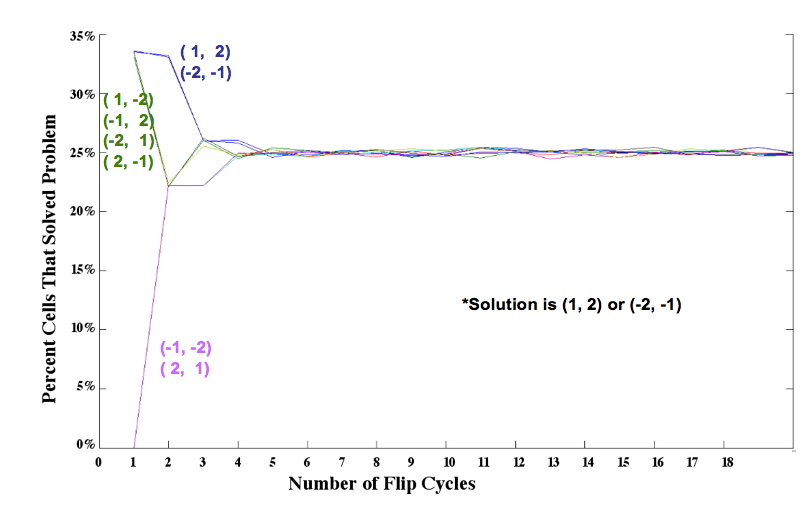 Simulation results for two pancakes, useful for calibrating kinetics of pancake flipping. |
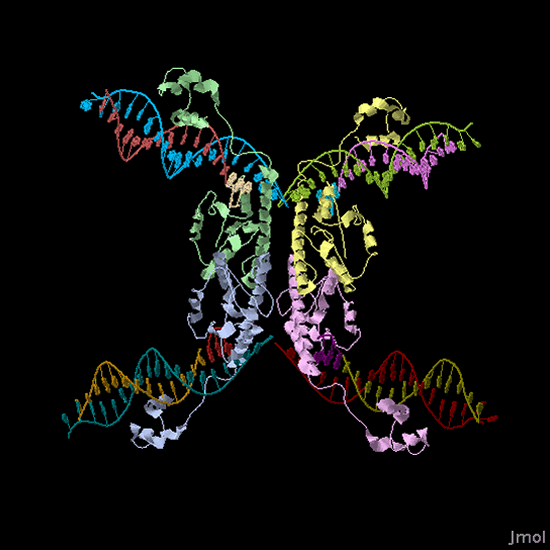 3-D structure of a Hin protein complex bound to DNA. View the interactive [http://www.rcsb.org/pdb/static.do?p=explorer/viewers/jmol.jsp Jmol image] (PDB 1ZR4). |
Methods and Results
Basic parts: Parts used in this project were designed by the [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006&group=Davidson Davidson] and [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006&group=Missouri Missouri Western] iGEM teams
Modeling
- Modeling the behavior of pancake flipping: deducing kinetics and size biases
- Using modeling to choose which families of unsolved pancake stacks to start with
Building the Biological System
- Single pancakes
- The problems of read-through - uncontrolled Tet expression, uncontrolled flipping
- New [http://partsregistry.org/Part:BBa_J31009 pSB1A7] vector: insulates, but is not compatible with parts carrying double terminators
- Designing pancakes without TT's
- Two pancake constructs
- Biological equivalence - distinguishing 1,2 from -2,-1 using RFP-RBS, updated panckaes
Conclusions
- Consequences of devices: data storage, possible application for rearranging transgenes in vivo, proof-of-concept for bacterial computers, first in vivo controlled flipping of DNA??
- Next steps: can solve problem but need control over kinetics
- Lessons learned:
- Troubleshooting, communication, teamwork, publicity
- Math and Biology meshed really well and even uncovered a new proof
- Multiple campuses can increase capacity through communication and cooperation
- Size of school is not a limiting factor
- We had a blast and learned heaps
TEAM MEMBERS
Students
- Sabriya Rosemond is a junior biology major at Hampton University.
- Erin Zwack is a junior biology major at Davidson College.
- Lance Harden is a sophomore math major at Davidson College.
- Samantha Simpson is a sophomore at Davidson College who might design a major in genomics.
Faculty
- A. Malcolm Campbell [http://www.bio.davidson.edu/campbell Department of Biology]
- Laurie J. Heyer [http://www.davidson.edu/math/heyer/ Department of Mathematics]
- Karmella Haynes [http://www.bio.davidson.edu/ Department of Biology - teaching postdoc and visiting assistant professor]
TOOLS AND RESOURCES
iGEM 2006 Jamboree
White Board
Biology Tools (Wet Bench)
Math Tools
- Pancake Simulators, aka The Lancelator (MATLAB Code)
- Other Flipping Simulators
- [http://gcat.davidson.edu/IGEM06/randompancakeflipper.m User-Friendly Version]
- [http://gcat.davidson.edu/IGEM06/powerflipper.m Power-User Version]
- [http://gcat.davidson.edu/IGEM06/powerflipperplus.m Power-User Update]
- [http://gcat.davidson.edu/IGEM06/findmin.m Findmin]
- [http://gcat.davidson.edu/IGEM06/bigtrial.m Bigtrial]
- Graphing Tools
- [http://gcat.davidson.edu/IGEM06/pancakeplotter.m Pancakeplotter]
- [http://gcat.davidson.edu/IGEM06/permplotter.m Permplotter]
- [http://gcat.davidson.edu/IGEM06/adjmat.m Adjmat - creates adjacency matrices]
- [http://gcat.davidson.edu/IGEM06/bioadjmat.m Bioadjmat - Adjmat with biological equivalence]
- [http://gcat.davidson.edu/IGEM06/make_radial.m Make_radial - creates a radially-embedded graph]
- [http://gcat.davidson.edu/IGEM06/find_diam.m Find_diam - finds the diameter of a pancake graph]
Bio-Math Tools
- [http://gcat.davidson.edu/IGEM06/signedperms.m Signedperms - lists all signed permutations for a stack of k pancakes]
- [http://gcat.davidson.edu/IGEM06/bioperms.m Bioperms - Signedperms with biological equivalence]
Assembly Plans
Progress
|