Harvard 2006
From 2006.igem.org
| Harvard University 2006 iGEM Team | ||
Students
|
Teaching Fellows
Faculty Advisors
| |
The Harvard University 2006 iGEM Team
This year Harvard's team consisted of 10 undergraduate students, with backgrounds in molecular and cellular biology, biochemistry, and computer science. With the help of five faculty advisors and three graduate-level teaching advisors, they devised and executed three separate projects.
1. Construction of novel DNA nanostructures for the purpose of stealth drug delivery,
2. Exploration of cell-surface targeting using interchangable and linkable aptamers (adaptamers) and Lpp-OmpA fusion vehicle for bacterial surface display.
3. Reconstitution of a circadian oscillator from cyanobacteria into E. coli,
DNA Nanobox: A Device for Stealth Drug Delivery
Project Overview
- We expect that molecular containers could have several interesting scientific and clinical applications, such as
- Drug and gene delivery
- Bio-marker scavenging (early detection of biomarkers for cancer or other progressive diseases)
- Directed evolution (compartmentalized selections)
- Using multiplexing for combinatorial chemical synthesis
- Capture and stabilization of multiprotein complexes
- Our goal for the summer was to design and implement nano-scale molecular containers, which could be dynamically opened and closed by an external stimulus. This is the first step to making more sophisticated drug-delivery vehicles.
- The containers were implemented as DNA nanostructures, which afford a significant degree of positional control and chemical versatility.
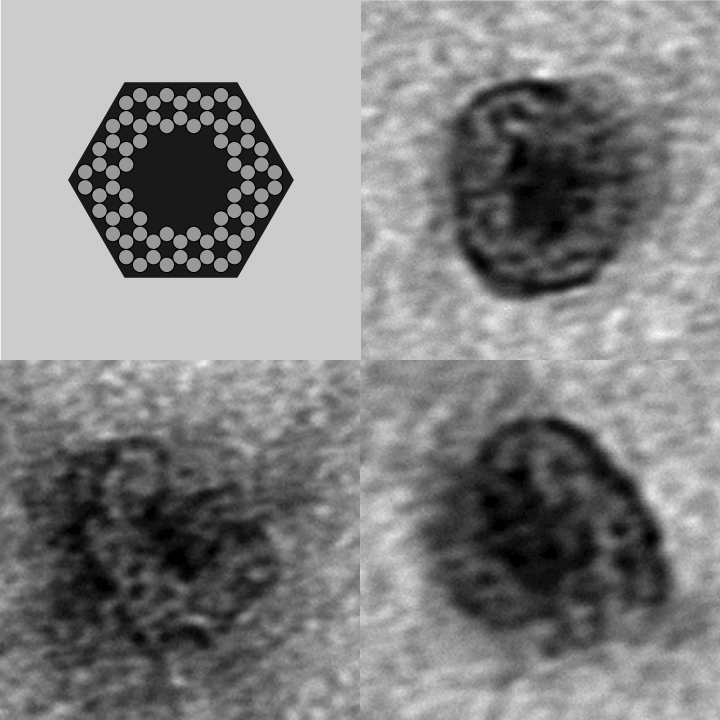
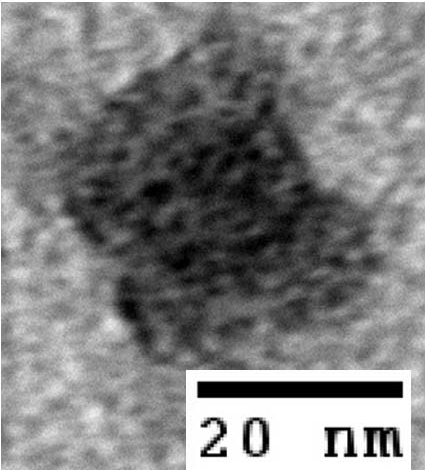
- To demonstrate that our designs successfully self-assembled, we used negative-stain electron microscopy to visualize the containers.
- As an initial proof-of-concept, we planned to demonstrate that our DNA containers could be used to "protect" biotinylated oligonucleotides from binding by streptavidin-coated magnetic beads.
- We were able to successfully show that nanoboxes with internal biotinylated oligonucleotides did not bind to streptavidin-coated magnetic beads, while nanoboxes that were externally decorated with biotinylated oligonucleotides were bound by the same beads.
Results
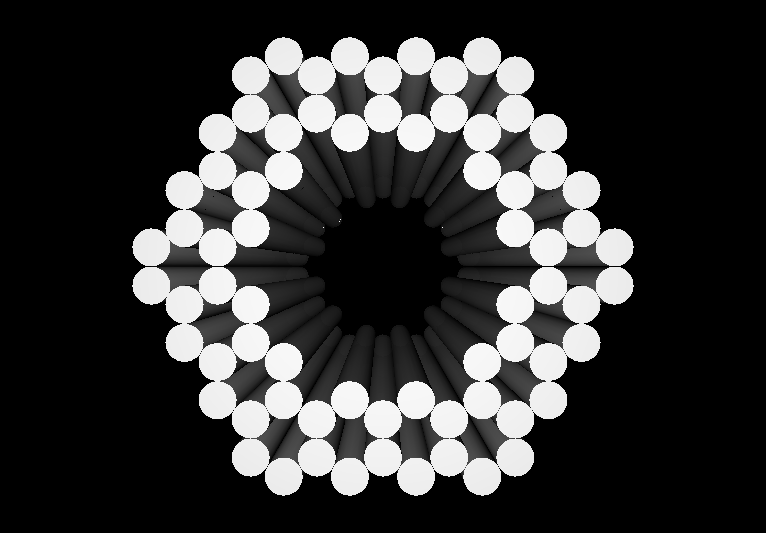
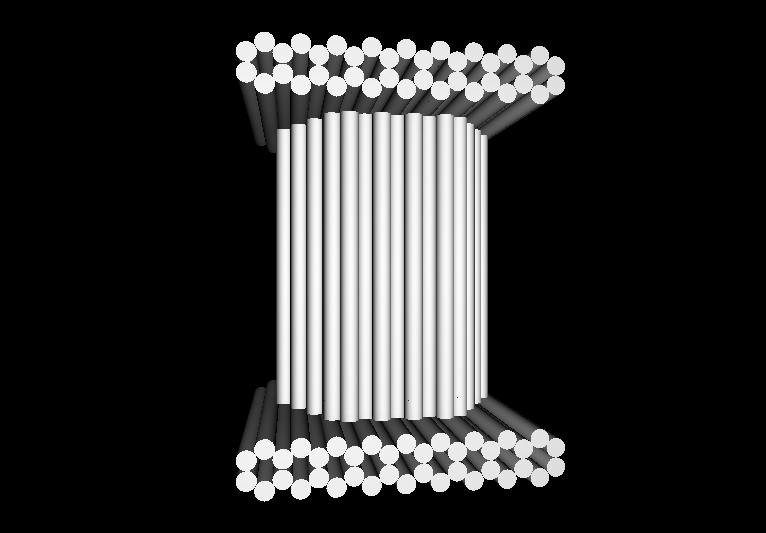
- The following images show our most successful container design, both as a 3D cartoon and in actual negative-stain EM images. The length, width, and depth of this container design is approximately 30x30x30 nanometers, so it is bigger than a typical ribosome.
- More details about this design can be found [http://openwetware.org/wiki/IGEM:Harvard/2006/Container_Design_5 here].
- The basic design of this container is a hexagonal barrel, closed off by two flat lids.
- The lids are designed to reversible attach to the barrel using complementary oligonucleotides using a "toehold" strategy in which the strands include unpaired overhangs that can be displaced with the addition of oligos that match for the full length of the sequence.
- Unfortunately, we did not have time to image our lid designs, or couple the lid and barrel and image the complete closed container.
Future Plans
- We have demonstrated the first steps toward building a sophisticated, versatile drug delivery vehicle out of DNA.
- Short term plans include assembly of closed containers (which include lids), and further characterization of assembled structures (both imaging and functional assays). We do not yet know what is the maximum size for a molecule that can still diffuse in and out of our container, but for a closed container we expect it to be around 1 nanometer in diameter.
- In the long term, we envision designing containers that incorporate cell-surface targeting mechanisms that would allow delivery of a payload to a specific cell type. Advances also need to be made if the container is going to deliver small molecules - perhaps by encapsulating the container with a liposome (or vice versa).
- [http://openwetware.org/wiki/IGEM:Harvard/2006/DNA_nanostructures openwetware page]
Cell Surface Targeting
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Cell_surface_targeting openwetware page]
Adaptamers
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Adaptamers openwetware page]
Project Overview
- We are interested in the mostly equivalent problems of targeting substrates to cells and cells to substrates. In the former case, targeting a substrate to the cell would facilitate uptake and a subsequent cellular response, important in fields such as drug delivery. Directing cells to particular places, for instance, a column, could be used to isolate cells and perform diagnostics.
- Briefly, we are pursuing two methods to attack this problem. In one route, we will express streptavidin on the E. coli cell surface, hence providing a target for any biotinylated molecule. In the second, we will build on the work of Tahiri-Alaoui et al. (2002) in developing bi-specific DNA "adaptamers" that can bind a cell surface and a substrate.
Results
Future Plans
Cell Surface Fusion Proteins
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Fusion_proteins openwetware page]
Project Overview
Results
Future Plans
A Circadian Oscillator for E.coli
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Cyanobacteria openwetware page]
Project Overview
Results
Future Plans
Complete Electronic Notebooks
Detailed records of our summer activities, including results, can be found on openwetware, which the team used to host its [http://openwetware.org/wiki/IGEM:Harvard/2006 electronic notebook].