X-Y chemotaxis
From 2006.igem.org
Contents |
Group I
Aim
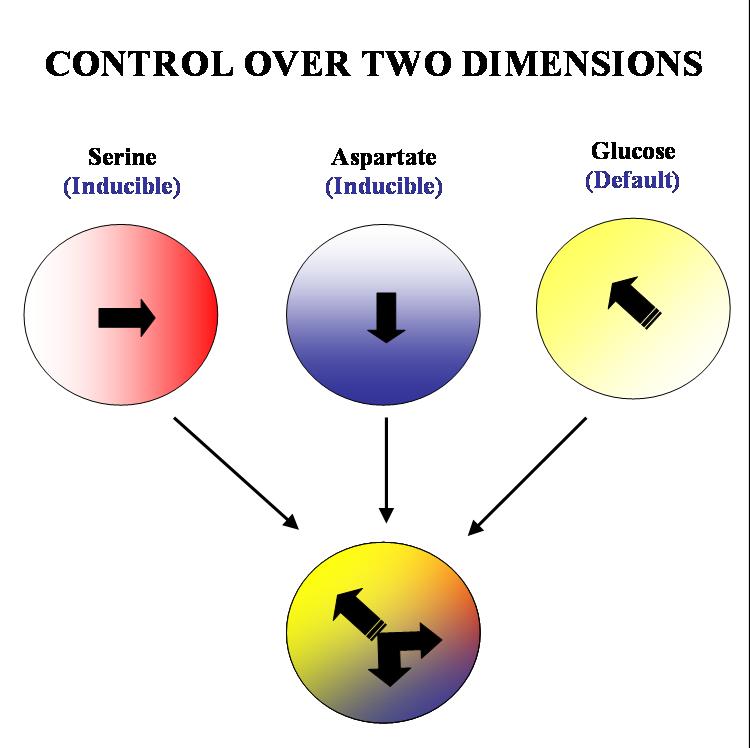
Our aim is to get complete two dimensional control over the movement of colonies of E. coli. on a solid substrate such as an agar plate. We plan to acheive this by 'hacking' into the existing chemotactic machinery in the E. coli cell. Specifically, if we are able to independently and efficiently alter the sensitivity of our bugs to stable orthogonal gradients of different amino acids, it will be possible to implement such control.
Approach
The specific geometry we have in mind is the following: there will be two stable gradients of serine and aspartate, orthogonal to each other. By inducing the expression of Tar and Tsr chemotaxis receptors (Tar for aspartate and Tsr for serine) in a null background (UU1250 strain) we hope to independently alter the response of the bug to these two gradients. There will also be a third gradient of glucose angled at 135 degrees to each of the amino acid gradients. Glucose is a PTS (phosphotransferase system) sugar and doesn't require Tar/Tsr receptor expression for its detection. Hence the bug will be constitutively responsive to glucose and this will serve as the default negative direction.
Constructs
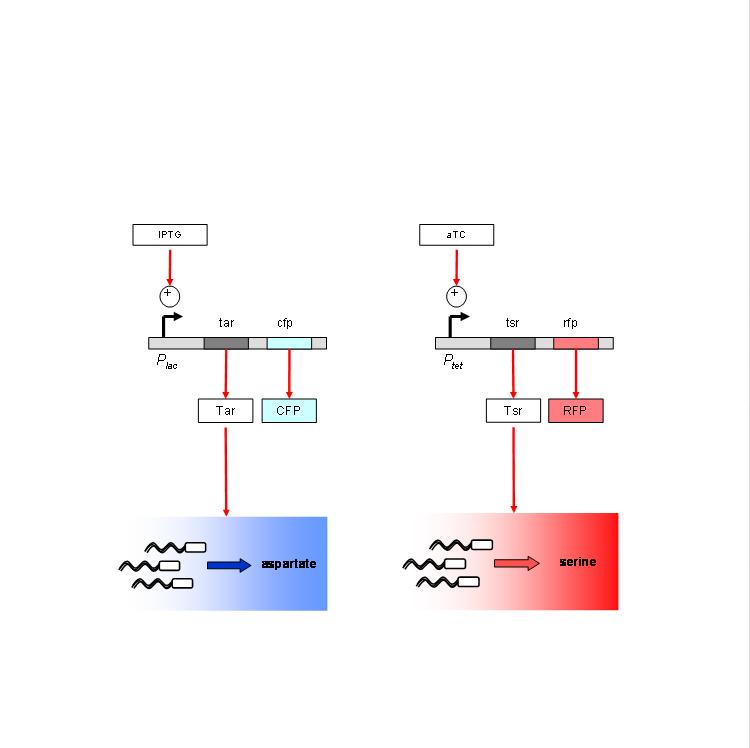
We are using the following constructs for induction of expression of Tar and Tsr. Tar expression is controlled by the lactose promoter, while Tsr expression is controlled by the Tet promoter. CFP and RFP provide readouts of relative transcript levels. We specifically chose these two reporters because of their spectral spacing in order to minimize bleed-through during fluorescence imaging.
The chemotaxis null strain, UU1250, first had to be transduced with chromosomal copies of the genes coding for repressor molecules for each of the promoters - lacI for the lac promoter, and TetR for the Tet promoter. This was done using P1 phage transduction with the donor strain K12zi. The selection marker was Spectinomycin. We selected 4 strains of the (lacI-TetR) transduced UU1250, named UU1250zi 4 through 7, and went ahead to test independent induction.
Induction
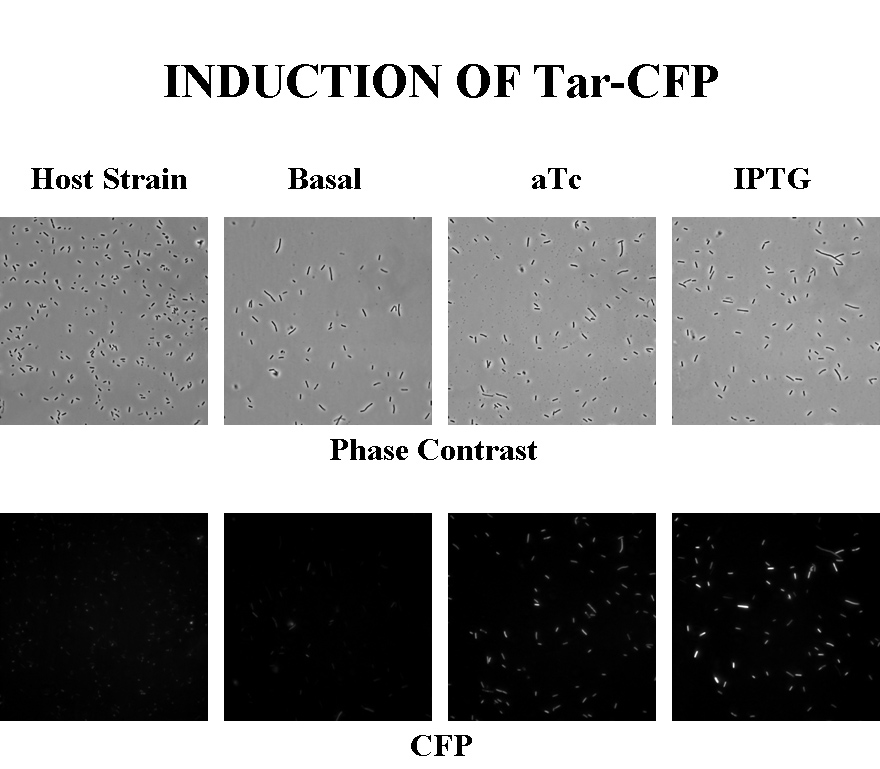
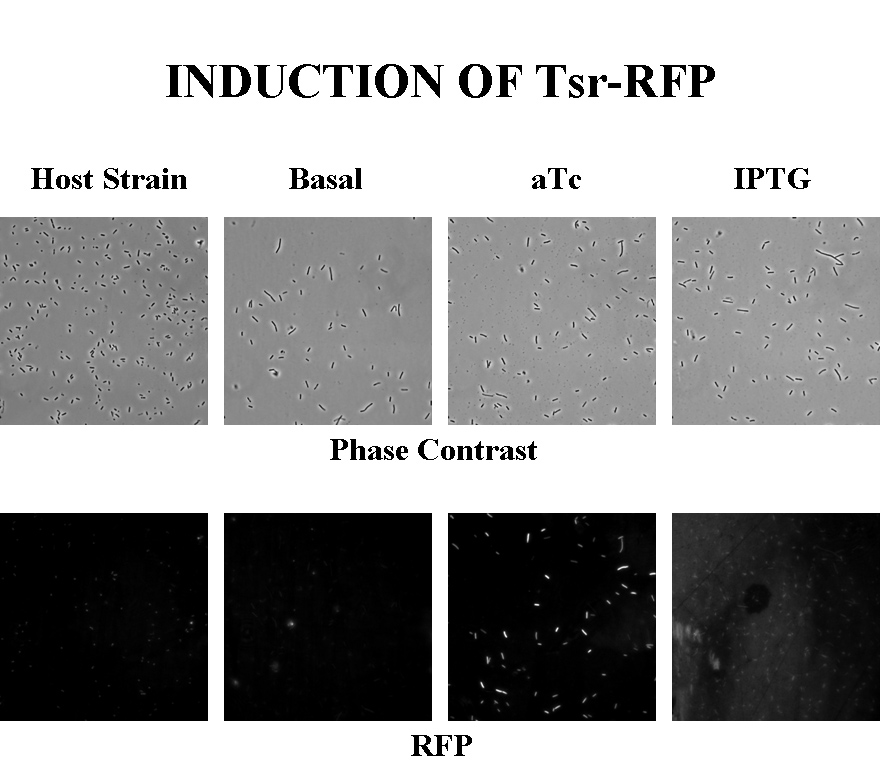
Briefly, UU1250zi, transformed with a plasmid containing both the Tar and Tsr constructs, was grown in LB broth + antibiotic overnight and then in succinate M9 minimal medium for a period of around 5 hours, with either IPTG or ATC (inducers for the lac and Tet operons respectively). A negative control to measure basal expression in absence of any inducer was included, while untransformed UU1250zi served as another negative control for autofluorescence levels.
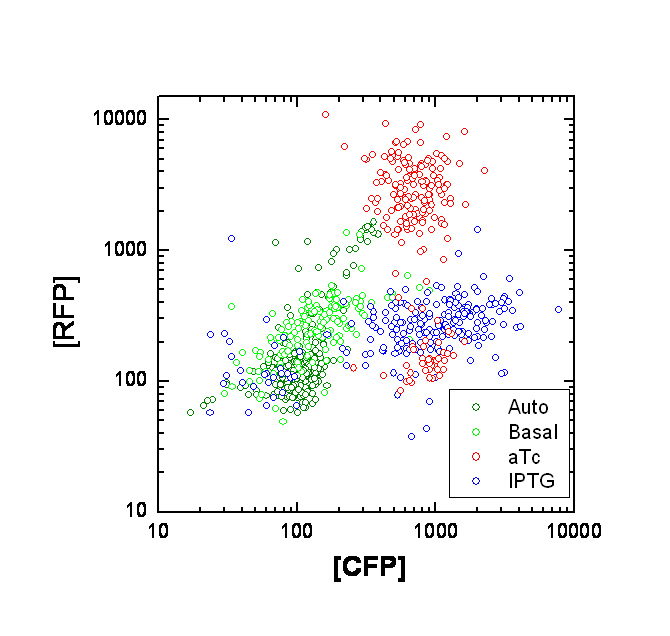
Each point on the scatter plot to the right indicates the total CFP v/s RFP fluorescence of a single E. coli cell, after correction for bleed-through and background subtraction. It is apparent that there is significant cross induction when ATc is added, since an increase in the CFP fluorescence also occurs.
A few explanations for the above anomaly :
If the Tsr-RFP construct was upstream of the Tar-CFP construct, it could be the case that the increase in CFP could be due to low efficiency of transcriptional termination, which would result in a long transcript containing both constructs. However this possibility is ruled out because the Tar-CFP construct is upstream in our case.
Alternately, this could be due to incorrect bleed-through correction. We need to confirm this with cells expressing CFP or RFP alone. A straightforward approach is to transform cells with only one of the constructs at a time.
We will also transform UU1250 (i.e. the original null strain without lacI and TetR repressors) with the constructs, to determine maximal expression levels, and to see where our induction stands on this scale.
Ruchi has done the next round of induction experiments. Results will be uploaded soon...
Attempts to establish such a gradient(s) using technigues standardised earlier include the slant plate. A KMnO4 gradient set-up is shown here: