Cambridge University 2006
From 2006.igem.org
| Line 57: | Line 57: | ||
*Las-sender (constitutive): BBa_I0407 | *Las-sender (constitutive): BBa_I0407 | ||
*Rhl-sender (constitutive): BBa_I0405 | *Rhl-sender (constitutive): BBa_I0405 | ||
| - | *Cin-sender (constitutive): BBa_I0409 <br> | + | *Cin-sender (constitutive): BBa_I0409 <br><br> |
''(c) AHL-induced fluorescence response:'' | ''(c) AHL-induced fluorescence response:'' | ||
*Lux-receiver (GFP): BBa_T9002 | *Lux-receiver (GFP): BBa_T9002 | ||
Revision as of 17:23, 24 October 2006
|
[http://www.ccbi.cam.ac.uk/iGEM2006 University of Cambridge iGEM 2006 - Complete Documentation] |
| - Autonomous Pattern Formation between Bacterial Populations by Reciprocal Communication - |
-
PROJECT SUMMARY
- ECFP: BBa_I13601
- GFP: BBa_J04430
- EYFP: BBa_I6031
- mRFP1: BBa_J04450
- Lux-sender (auto-inducing): BBa_I15030
- Las-sender (constitutive): BBa_I0407
- Rhl-sender (constitutive): BBa_I0405
- Cin-sender (constitutive): BBa_I0409
- Lux-receiver (GFP): BBa_T9002
- Lux-receiver (EYFP): BBa_I13263
- Las-receiver (EYFP): BBa_I0426
- Rhl-receiver (EYFP): BBa_I0424
- Cin-receiver (EYFP): BBa_I0428 <p>
- pSB1A2:BBa_P1010
- pSB1A3:BBa_P1010
- pSB1AC3:BBa_S03594
- pSB1AC3:BBa_S03541
- pSB1AC3:BBa_S03549
- pSB1AC3:BBa_S03550
- Las Autoinducer Cassette (can also be used in 3A Assembly): BBa_J28029
- Gatsby Foundation
- DNA 2.0 Inc.
Our entry for the iGEM 2006 competition sought to genetically engineer two types of bacteria such that they would autonomously form patterns in swimming agar. We studied the dynamics of pattern formation between bacterial populations. In particular, we investigated the interaction of genetically engineered E. coli sender cells, producing an acyl homoserine lactone signal, and receiver cells in swimming agar. We then attempted to render these cells capable of reciprocal communication and sought to construct biological parts that would render possible the construction of a population based bi-stable switch. Furthermore, we modelled the behaviour of single cells as well as the interactive behaviour of populations of cells containing these genetic circuits. Our experiments verified that differential cell motility in combination with position-dependent gene expression has the potential to generate complex patterns.
INTRODUCTION
Position dependent gene expression is a critical aspect of the behaviour of multi-cellular organisms and requires a complex series of interactions to occur between cell types. We used Escherichia coli as the organism of choice for our experiments and observed how differential cell motility can in itself lead to complex pattern formation (Fig 1.1). We genetically engineered an E. coli cell population to render it capable of producing an acyl-homoserine lactone signal (sender cells). We verified that the cells were producing lactones using the CVO26 plate assay described by McClean et al. (Fig 1.2). A second E. coli population was rendered capable of responding to this acyl homoserine lactone signal (receiver cells). Adapting the experiments of Weiss et al., using cell motility, rather than a differential response to AHL concentrations as a way to define zones of response we noted how the interaction of sender and receiver cell populations on a swimming plate leads to complex pattern formation (Fig 1.3). For both experiments mentioned above we used parts from the MIT Registry of Standard Biological Parts, thus characterizing their function in this novel fashion. Thereafter, we proceeded to make the genetic constructs described in “Biological Parts” below. Equipping highly motile strains such as E. coli MC1000 with AHL-mediated autoinducing systems based on Vibrio fischeri luxI/luxR and Pseudomonas aeruginosa lasI/lasR cassettes would allow the amplification of a response to an AHL signal and its propagation. Exploiting the uncoupled nature of sender and receiver entities, both AHL systems can be combined to give two-way communication pathways. The recursive behaviour of cells being both sender and receiver at the same time has the potential to create patterns that are robust and more complex in nature. Although we were unable to construct the complete system, we were able to construct auto-inducing luxI/R and lasI/R cassettes. For some of our constructions we used the method of 3-Antibiotic assembly, described by Tom Knight et al. We were able to purify standard plasmid backbone vectors for this assembly method and also to get it to work for some of the constructions as evidenced by the parts submitted. We also managed to model the behaviour of cells within populations containing these systems. Further, we believe that in the future it may be possible to equip E. coli populations with more complex genetic circuits that would allow two interspersed subpopulations of isogenic bacteria to battle for dominance on a swimming plate. This is described in detail in "Future Directions".
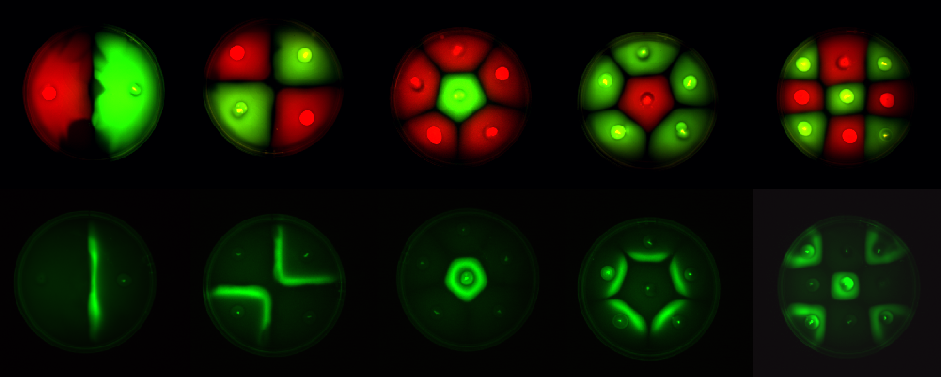
Fig1.3: All inoculations were with E. coli MC1000. The constitutive red fluorescence is with BBa_J04450; that of green fluorescence is with BBa_J04430. The sender/receiver assay in the bottom panel is based on lux-AHL signalling. Sender: BBa_I15030 Receiver: BBa_T9002 |
|
| ||||||||
BIOLOGICAL PARTS
1. Parts used for generating patterns (these are parts whose function we characterized)
(a) Constitutively expressed fluorescent proteins:
Cell Type 1

Cell Type 2

3. Parts submitted: these (except for BBa_p1010 PLASMIDS) could be used for system described in the 'Future Direction'
EXPERIMENTS & RESULTS
Aside from constructing genetic circuits, we conducted a number of experiments with bacteria equipped with these genetic circuits. Broadly speaking we conducted four types of experiments, each of which is described below:
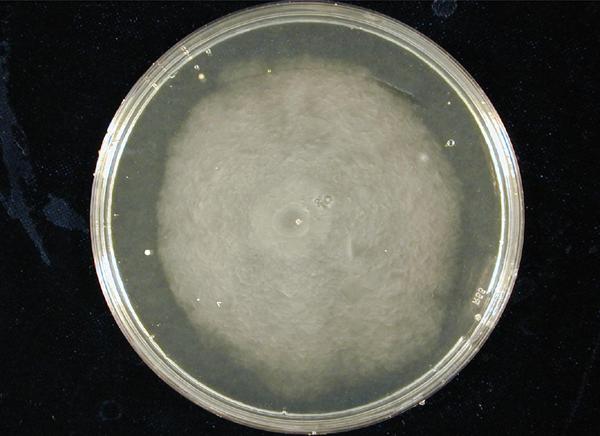
1. Standardization of the swimming assay for different strains of E. coli and using differential cell motility for pattern generation: We were able to standardize the swimming assay for E. coli and established conditions under which E. coli cells would swim when a swimming agar plate was innoculated with an E. coli cell population. We found that E. coli cells would swim only in agar medium containing 0.3% bactoagar, 0.5% NaCl and 1% Bactotryptone. Further, optimum swimming was observed on incubating the plates at 30 C overnight and pouring the agar at an optimum temperature. (Fig 3.1 depicts a swimming assay) To further examine swimming behaviour we prepared microscale agar plates, innoculated them with E. coli (MC-1000) expressing GFP and observed these under a wide-field inverted microscope. After standardizing the conditions for swimming we innoculated agar plates with different strains of E. coli to determine which of them was the most motile. We used strains MC-1000, MG-1655, MC-1000 FlhD-Kan and XL-1 Blue. We determined that MC-1000 was the most motile strain, followed by MG-1655 and XL-1 Blue while the mutated version of MC-1000 was not motile. Interestingly, we observed that if a swimming agar plate is innoculated with two different populations of E. coli, each being differently motile, this allows the generation of patterns (Fig 3.2). It was also interesting to note a clear zone at the interface between two different bacterial populations swimming from opposite ends of the plate as would be evident from all our photos. We can only hypothesize about the reason for this. To further examine why this is so, we used a wide-field inverted microscope again and noted that the clear zone contains no bacteria. This is possibly a result of nutrient depletion in that region.
2. Transforming motile strains of E. coli with fluorescent proteins and observing the patterns generated on swimming agar plates by such strains: We transformed different strains of E. coli with genetic circuits from the Registry of Biological Parts, such that they would express fluorescent proteins. We thus generated strains expressing green Fluorescent protein, cyan Fluorescent protein, yellow fluorescent protein and red fluorescent protein. We then used these fluorescent strains to generate vivid patterns. We found that experimentally it was easiest to distinguish green fluorescent strains from red fluorescent strains as these emit light in distinct spectral regions.
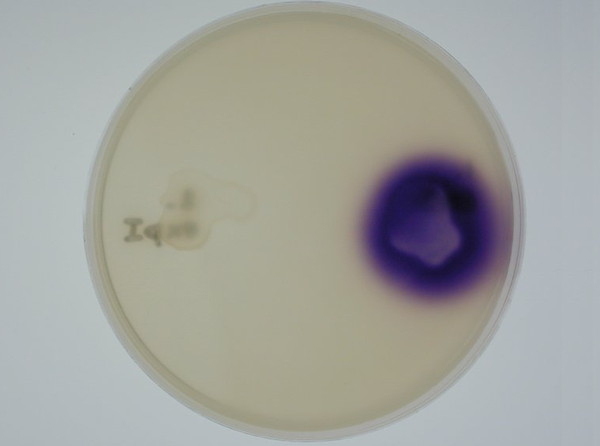
3. Equipping E. coli with biological parts and rendering them capable of acyl homoserine lactone (AHL) production and assaying this using a CVO 26 assay: We equipped E. coli populations with biological parts (described above) that rendered them capable of producing acyl homoserine lactones. We confirmed that these E. coli strains were indeed producing lactones by using a mutated strain of Chromobacterium violaceum CVO26 and using the assay described by McClean et al (of Nottingham State University). We further found that violacein production by CVO 26 on exposure to lactones could itself be used to generate interesting patterns (Fig 3.3).
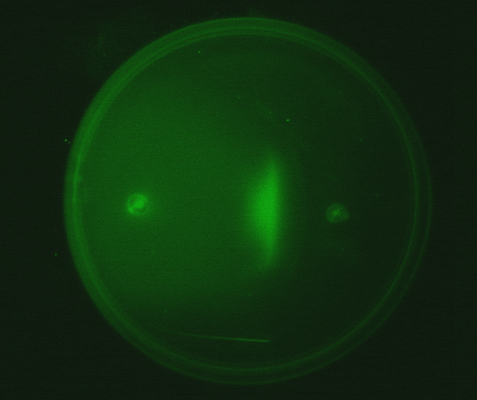
4. Observing the interaction of two E. coli populations one producing AHL and the other capable of responding to it: We equipped a second E. coli population with biological parts (described above) that rendered it capable of responding to AHL by expressing green fluorescent proteins. We then observed the interaction of this population with the population described in (3) in swimming agar. We observed that this interaction can lead to very interesting patterns, with differential cell motility defining zones of response (Fig 3.4).
|
| ||||||||
MODELLING
Prior to any 'wet' work, we sought to build mathematical models for our genetic system. We used these models to verify whether in theory the system is likely to work and if so, how it would behave. The single-cell model describes the dynamics inside a cell, whereas the multi-cell model simulates the interaction between cells on a global scale.
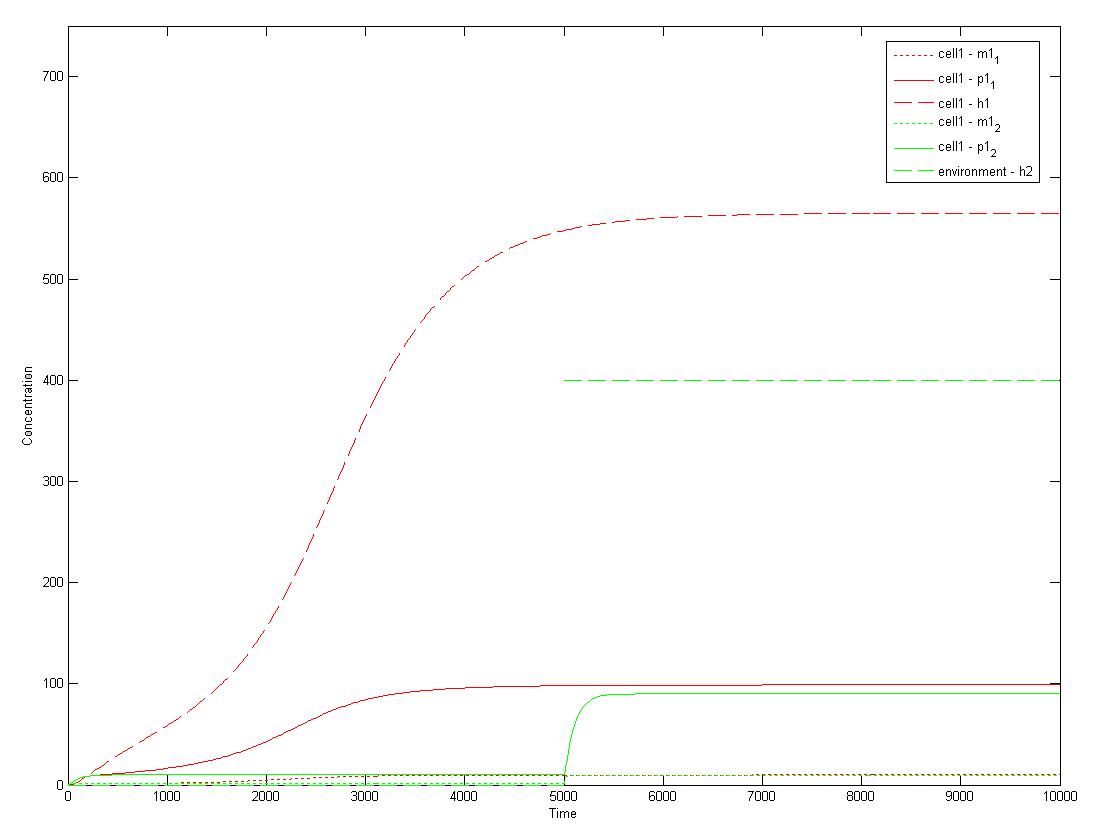
1. Single-cell mathematical model
This is a Matlab model of the dynamic behaviour of Type 1 Cell (Fig 4.1). Note that although there are two types of cells in our system, their dynamics should be similar and here we only modelled Type 1 Cell, i.e. it constitutively expresses mRFP (Red) and starts to express GFP (Green) on recieving AHLs from Type 2 Cells. On the diagram, the dashed lines refer to the concentration of AHLs; dotted lines the concentration of mRNAs; solid lines the concentration of proteins (individual cistrons are not distinguished). As shown in Fig 4.1, the auto-induction is indicated by an initial exponential increase in protein production and then a tail-off to its limiting concentration (solid red line). The diagram also shows a switch and increase in GFP expression as the cell recieves its opponent AHLs (solid green line 'switches on' after a step input of AHLs indicated by the dashed green line).
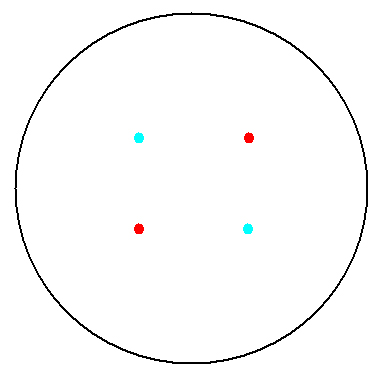
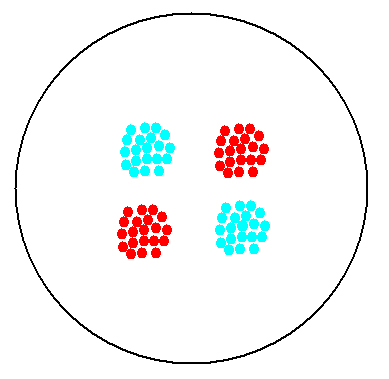
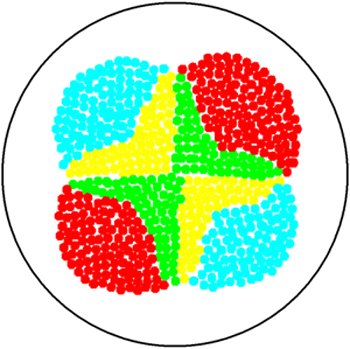
2. Multicellular Interaction Computer Simulation
This is essentially a computer simulation program that simulates multi-cellular interactions on a swimming plate. The diagrams below (Fig 4.2 - 4.4) illustrate these: initially we have four innoculations of two types of cells (e.g. same types are innoculated diagonally); the cells then swim and replicate over a certain time period, thus growing larger in number; eventually due to AHL diffusion and cell motion, the cells respond to their opponents and start to change colours, i.e. Cyan turns to Yellow, Red turns to Green, giving a cross pattern (in the end we would expect the plate to be full of Yellows and Greens, instead of the initial Cyans and Reds). The simulation program shows that various patterns could be generated via different innoculations, just as those shown in Fig 1.3.
|
| ||||||||
APPLICATIONS
The experiments we conducted and the results we obtained shed some light on how position dependent gene expression may occur during the development of multi-cellular organisms. Such experiments also shed light on tissue invasion and metastasis. Metastasizing cells break away from a primary tumour, penetrate into lymphatic and blood vessels and then colonize new regions of the body. Metastasizing cells are further characterized by the fact that they express unique genes not expressed by normal cells. Tissue invasion requires the movement of cells. Eukaryotic cell movement is far more complex than prokaryotic cell movement but it is of interest to observe the factors regulating prokaryotic cell movement and how swimming cells may express genes in only certain locations when exposed to a signal gradient. Biofilm formation also requires cell movement and position dependent gene expression. Further, lactone signalling is involved in biofilm formation of Pseudomonas aeruginosa. It would be very interesting to study the interaction of bacterial populations capable of two-way communication as many complex processes in higher organisms involve two way communication. The construction of a population-based bistable switch would shed even more light on these processes.
FUTURE DIRECTION
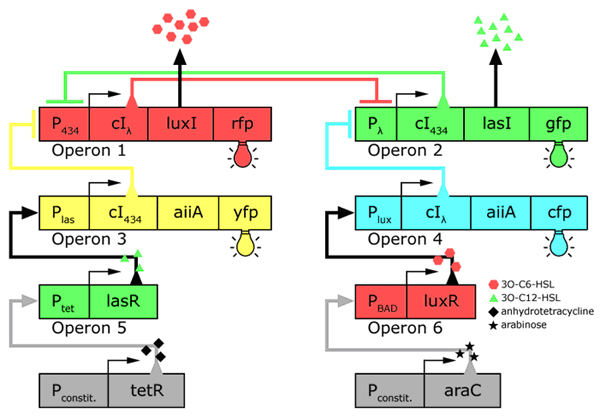
In the future it may be possible to equip E. coli populations with more complex genetic circuits that would allow two interspersed subpopulations of isogenic bacteria to battle for dominance on a swimming plate. They would initially be either red or green; on encountering each other in sufficient numbers, defeated bacteria would defect to the opposing side by a switch mechanism. They would change colour and start producing a new battle signal to command the fight under the opposite banner. An overview of the genetic circuitry is given below: there are two competing gene expression states, illustrated with red and green fluorescent proteins, i.e. rfp and gfp gene products. The ratio of expression from operons 1 and 2 represents the state of a cell. Operons 3 and 4 induce switching of the cell's state or "defection" in the face of an overwhelming opposite-state majority (sensed by HSL signals). If the opposite HSL battle signal is above a certain threshold, an overwhelmed cell switches gene expression and defect to the other state. We have submitted some parts to the Registry that would make easier the engineering of these complex genetic circuits and the construction of such a population based bi-stable switch a more realistic goal.
|
Genetic circuitry of the future system |
THE TEAM
Students
Elizabeth Carter | Jisun Lee | Kaj Bernhardt | Nikhilesh Chand | Xueni Zhu | Yang Xu
Faculty
Gos Micklem | Jim Ajioka | Jim Haseloff | Jorge Goncalves
Contributors:
Gillian Fraser | Glenn Vinnicombe | James Brown | Jan Lowe | Jason Chin | Keith Johnstone | Matthew Levin | Pentau Liu | Rita Monson
ACKNOWLEDGEMENTS