Event Processing Device
From 2006.igem.org
(→Input-module Schematic) |
(→Input-module Development) |
||
| Line 37: | Line 37: | ||
¦--------------------------------------------------> PoPS_in, t | ¦--------------------------------------------------> PoPS_in, t | ||
0 1 | 0 1 | ||
| - | |||
| - | |||
| - | |||
==Input-module Schematic== | ==Input-module Schematic== | ||
| Line 48: | Line 45: | ||
legend: | legend: | ||
| - | |||
I: input signal that will bind to the promotor P_in, e.g. heat shock dependent etc. | I: input signal that will bind to the promotor P_in, e.g. heat shock dependent etc. | ||
P_in: promotor that allows the signal of choice I to bind. | P_in: promotor that allows the signal of choice I to bind. | ||
| Line 63: | Line 59: | ||
R_n: toggle switch gene(s) | R_n: toggle switch gene(s) | ||
| - | == | + | Any input can be used, as long as a romoter can be found that is either activated or suppressed by it. At the output any kind of genes can be added which will be produced depending on the PoPS of that particular input. |
| - | + | ||
| - | [ | + | |
| - | + | ==Suitable Strategies== | |
| + | As shown in the black box above, the core of the device is a regulator sub-module, for which [[INPUT-module_regulator | different implementation strategies]] have been investigated. | ||
| + | |||
| + | We identified various solutions, the remaining ones being | ||
| + | |||
| + | * The λ-system with two variants | ||
| + | ** The original anti-parallel version | ||
| + | ** An engineered unidirectional version | ||
| + | * The Lux-system | ||
| + | |||
| - | |||
| - | |||
| - | |||
| - | |||
'''To be continued: [http://www.ek.ethz.ch/iGEM/Input-module.planning.050905.doc input-module docu as word document] for now (maintained by dominic). Details of the literature research on the two strategies above should be extended by Christophe and Hervé themselves''' | '''To be continued: [http://www.ek.ethz.ch/iGEM/Input-module.planning.050905.doc input-module docu as word document] for now (maintained by dominic). Details of the literature research on the two strategies above should be extended by Christophe and Hervé themselves''' | ||
Revision as of 08:09, 19 September 2005
Contents |
Organisational
Group Members
- Christophe, Dominic (coordinator), Giorgia, Herve, Zlatko
Group Meeting History
log 2005-08-17: Wednesday, 13:00 @ polyterrasse: Discussion of module, next steps, task distrib. log 2005-08-22: Monday, 15:00 @ polyterrasse: Discussion of biol. solutions. log 2005-09-12: Thursday, 17:30 @ teammeeting.
Input-module Development
Introduction
The iGEM-team at ETH decided to implement a modular and standardized Counter-system that can in principle be used to count any kind of biological event and generate any kind of output after a certain number N of events has been detected. The Counter-system can be split into 2 basic units, the NOR-module and the INPUT-module (outlined in red in the schematic below). This document deals with the implementation of the INPUT-module.
Module Description
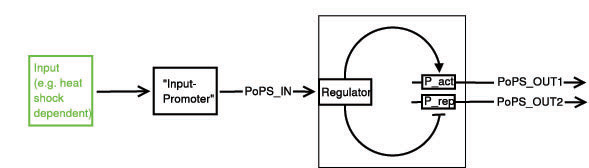
In a nutshell, the Input module has 2 system boundaries, both of which are characterized by PoPS (Polymerase Per Second). Its purpose is to take a single input PoPS and output 2 different PoPS. One of the outputs should be high and the other low when S is high and vice versa when S is low.
PoPS_outn
^
¦
high ¦ PoPS_out2 _______ _________ PoPS_out1
¦ \ /
¦ \ /
¦ \ /
¦ \ /
¦ \ /
¦ X
¦ / \
¦ / \
¦ / \
¦ / \
low ¦ PoPS_out1 _______/ \_________ PoPS_out2
¦--------------------------------------------------> PoPS_in, t
0 1
Input-module Schematic
Below a preliminary parts-view of the module, i.e. encapsulation of biological specific implementations into a functional box with general PoPS interfaces.
legend: I: input signal that will bind to the promotor P_in, e.g. heat shock dependent etc. P_in: promotor that allows the signal of choice I to bind. PoPS_in: polymerase per second dependent on promotor and concentration of I Reg: regulation genes. there are different solutions possible, see below. r: repressing signal. highly dependent on Reg, P_r and of course speed and binding considerations a: activating signal. highly dependent on Reg, P_a and of course speed and binding considerations rep: repression, indicated by horizontal bar act: activation, indicated by arrow P_r: promotor region to be repressed and/or "roadblock" region. constitutively active P_a: promotor region to be activated and/or "roadblock" region. not constitutively active PoPS_out1: polymerase per second dependent on P_r and r. PoPS_out2: polymerase per second dependent on P_a and a. R_n: toggle switch gene(s)
Any input can be used, as long as a romoter can be found that is either activated or suppressed by it. At the output any kind of genes can be added which will be produced depending on the PoPS of that particular input.
Suitable Strategies
As shown in the black box above, the core of the device is a regulator sub-module, for which different implementation strategies have been investigated.
We identified various solutions, the remaining ones being
- The λ-system with two variants
- The original anti-parallel version
- An engineered unidirectional version
- The Lux-system
To be continued: [http://www.ek.ethz.ch/iGEM/Input-module.planning.050905.doc input-module docu as word document] for now (maintained by dominic). Details of the literature research on the two strategies above should be extended by Christophe and Hervé themselves