IPN UNAM 2006
From 2006.igem.org

Contents |
First bulletin of iGEM MEXICO November 2006 (bulletin in Spanish). Media:Bulletin-1.pdf
[http://www.ipn.mx/ Instituto Politécnico Nacional-Universidad Nacional Autónoma de México, México]
Mexican Staff - iGEM MEXICO
| STUDENTS | INSTRUCTORS | ||
| María E. González Jiménez maruhtfl#hotmail.com | Genaro Juárez Martínez (COM) genarojm#correo.unam.mx | ||
| Raúl I. López Gómez raul.ivan.lopez.gomez@hotmail.com | Pablo Padilla (COM) pablo#mym.iimas.unam.mx | ||
| Tania G. Bermúdez Cisneros tgbermudez#hotmail.com | Edgar Salgado Majarrez (BIO) esalgado@ipn.mx | ||
| Paulina A. Leon Hernández pauanana#gmail.com | Juan S. Aranda Barradas (BIO) jaranda@acei.upibi.ipn.mx | ||
| Pablo Gerardo Padilla tyomero#gmail.com | Ma Carmen Oliver Salvador(BIO) oliveripn@hotmail.com | ||
| Izchel N. Ríos Gaspar nayb_juice#yahoo.com.mx | Paola B. Zarate Segura (BIO) Pbzars@yahoo.com | ||
| Arturo Rodríguez Martínez arturo_rguez_mtz#yahoo.com.mx | Claudia I. Franco Arteaga (BIO) claudia_imelda@yahoo.com | ||
| Julio N. Argota Quiróz julioargota#hotmail.com | Rosaura Palma Orozco (COM) rpalma#math.cinvestav.mx | ||
| Alejandra Sánchez Arzate alesa#ciencias.unam.mx | Arturo Becerra Bracho (BIO) abb#fciencias.unam.mx | ||
| Iván Y. Fernández Rosales najtaaj#gmail.com | Elias Samra Hassan (COM) elias#uxmcc2.iimas.unam.mx | ||
| José C. Rodríguez Chico rodriguez_chico#hotmail.com | Carlos Silva Sánchez (COM) sscarlos#gmail.com | ||
| Rogelio Basurto Flores larckov#hotmail.com | Jaime López Rabadan (COM) rabadanlorj#gmail.com | ||
| Rosario E. Gordillo Padilla extraterrestre710#hotmail.com | Claudia Benítez (BIO) beni1972uk#yahoo.com.mx | ||
| Juan C. Gómez Sánchez thunder2099#hotmail.com | |||
| Armando Galicia Naranjo armandushko#hotmail.com | |||
| Abraham J. Leal Baena fenixaj#hotmail.com |
BIO: Biology, COM: Computation, PHY: Physics
Line of Investigation
Our main line of investigation uses unconventional computing as a means of building novel computing paradigms, such as in biology, physics and chemistry.
We are attempting to construct discrete dynamical systems capable of simulating cellular computation. Therefore, our work focuses on molecular computation. We thus have three models using cellular automata theory (you can see our original contributions [http://uncomp.uwe.ac.uk/genaro/ click here]).
Cellular automata (CA) operate over a scales ranging from the molecular level to n-dimensions and are massively parallel. Also, they are capable of supporting universal computation, self-repair and self-reproduction.
We wish contribute to the iGEM project development various protein based bio-components. We will work along three main lines: complex and reversible dynamical systems and formal languages, that support particles and multiple reactions, related to the molecular transformations.
We aim to develop CA software for a specific bio-simulation and gradually implement broader real biological scenarios.
iGEM México Project
EXPERIMENT PROPOSAL: Engineering a genetic signaling cascade to produce a green flourescence protein expression/repression system in Escherichia coli.
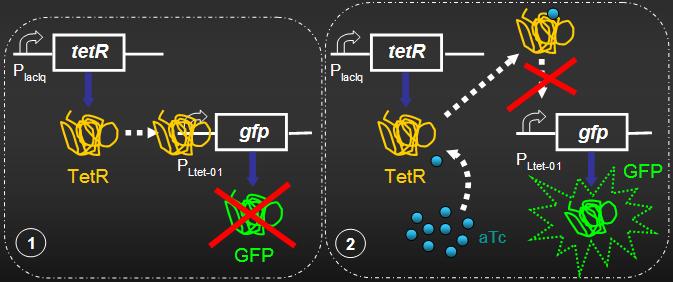
IN THIS PART OF THE PROJECT: We intend to emulate some genetic networks already identified in Arabidopsis responsible for the formation of hair in root and leaves. These networks lead to simple genetic circuits of the repression/expression type. We would like to show that these systems support Turing patterns.
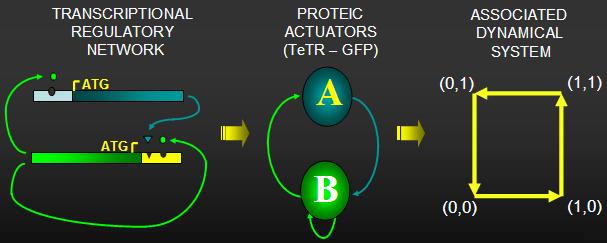
MODELLING PROPOSAL: Developing models to describe inside-the-cell metabolic events
through a cellular automata approach.
We study two-dimensional cellular automaton, where every cell takes states 0 and 1 and updates its state depending on sum of states of its 8 closest neighbors as follows. Cell in state 0 takes state 1 if there are exactly two neighbors in state 1, otherwise the cell remains in state
0. Cell in state 1 remains in state 1 if there are exactly seven neighbors in state 1, otherwise the cell switches to state 0. CA governed by such cell-state transition rule exhibits reaction-diffusion like pattern dynamics, so we call this Diffusion Rule.
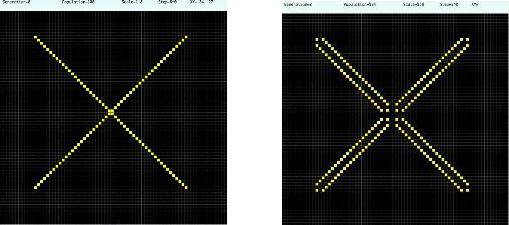
Using the diffusion rule we can generate a dynamical pattern over a system, like turn on/off
ligth with alive o dead cells that shows a luminescence, examples include fluorescence,
bioluminescence and phosphorescence.
Starting with any configuration, the cells alive are represented in yellow (the activator) and dead in black (the inhibitor), see figure 4. The system is created defining an inicial state over the base configuration (see figure 3). The luminescence is obtained by the evolution of this initial pattern.

Collaborators
Paula Figueroa Arredondo (BIO), Absalom Zamorano (BIO), Jovita Martínez (BIO), Juan C. Seck Tuoh Mora (COM), Sergio V. Chapa Vergara (COM), Francisco Hernández Quiroz (COM), Carlos A. Espinosa Soto (BIO), Mark Olson (BIO), Rafael Baquero (PHY), José U. Cruz Cedillo (COM), Rafael Peña Miller (COM), Rocío Reséndiz Muñoz (COM) and Ulises Vélez Saldaña (COM)
Institutions
 Instituto Politécnico Nacional (National Polytechnic Institute)
Instituto Politécnico Nacional (National Polytechnic Institute)
 Universidad Nacional Autónoma de México (National Autonomous University of Mexico)
Universidad Nacional Autónoma de México (National Autonomous University of Mexico)
 Universidad Autónoma del Estado de Hidalgo (National Autonomous University of Hidalgo State)
Universidad Autónoma del Estado de Hidalgo (National Autonomous University of Hidalgo State)
