Imperial College 2006/Sandbox
From 2006.igem.org
(→Our Open Documentation) |
(→Main Project) |
||
| Line 40: | Line 40: | ||
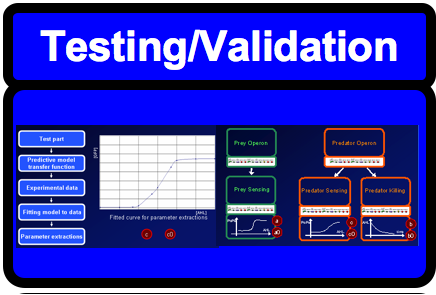
*Testing of different components of the oscillator, thus characterizing the parts | *Testing of different components of the oscillator, thus characterizing the parts | ||
| align="right"|[[Image:ArrowCurvedRight.png|110px]] | | align="right"|[[Image:ArrowCurvedRight.png|110px]] | ||
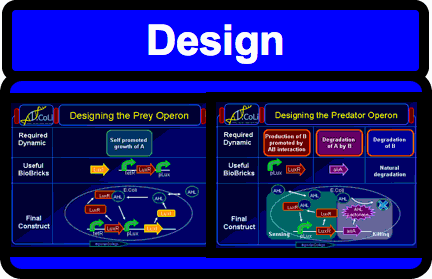
| - | | align="center" |{{Click || image=IGEM_IMPERIAL_DevCycle_Design.png | link=IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System/Design | width=200px| height=130px}} | + | | align="center" |{{Click || image=IGEM_IMPERIAL_DevCycle_Design.png | link = http://openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Oscillator/project_browser/Full_System/Design | width=200px| height=130px}} |
| align="left"|[[Image:ArrowCurvedDown.PNG|120px]] | | align="left"|[[Image:ArrowCurvedDown.PNG|120px]] | ||
|- | |- | ||
Revision as of 01:46, 30 October 2006
Imperial College London iGEM 2006
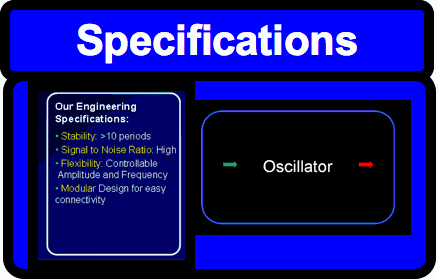
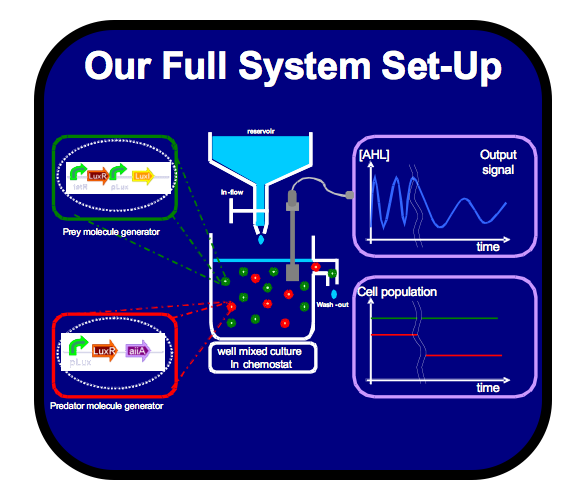
| Oscillators are a fundamental building block in many engineering fields and are a widespread phenomenon in biology. Building a biological oscillator is thus a crucial step forward in the field of Synthetic Biology. | Following the design of the oscillator, a full theoretical analysis of the Lotka-Volterra properties was carried out, which promised the successful outcome of oscillations. Additionally, all components as well as the overall oscillator were modelled such that the behaviour of the system could be predicted. Our team successfully built functional parts, thus providing the building block for the final oscillator. All parts created were experimentally tested and their characterization could be used to feedback information into the modelling. |
Main Project
Secondary Projects
| [http://openwetware.org/wiki/IGEM:IMPERIAL/2006/project/popsblocker PoPs Blocker] | [http://openwetware.org/wiki/IGEM:IMPERIAL/2006/project/Bio_elec_interface Biological to Electrical Interface] |
|---|---|
|
As a method of controlling the activation of the positive-feedback loop in our predator-prey based oscillator, we successfully created this part, which can be used as a general Pops Blocker:
|
We also worked on a Biosensor for measuring AHL concentrations in order to establish a Biological to Electrical Interface this summer.
|
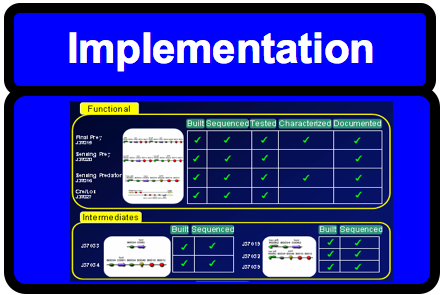
Our Contributions to the Registry
| Part Logo | Description | Link to registry | Built | Tested | Characterized | Sequenced | Sent |
|---|---|---|---|---|---|---|---|
| Final Constructs | |||||||
| Final Prey Cell | J37015 | YES | YES | Pending | YES | YES | |
| | Cre/Lox Prey Control | J37027 | YES | YES | YES | YES | YES |
| Test Constructs | |||||||
| Final Polycistronic Predator Cell Test Construct | J37016 | YES | YES | YES | YES | YES | |
| | Predator Cell, pLux Transfer Function (two promoters) | J37020 | YES | YES | NO | NO | YES |
| Intermediate Parts | |||||||
| | AHL induced LuxR generator (for predator cell) | J37019 | YES | N/A | N/A | YES | YES |
| | RBS + LuxR | J37033 | YES | N/A | N/A | YES | YES |
| | Prey Cell Intermediate | J37034 | YES | N/A | N/A | YES | YES |
| | LuxR + GFP | J37032 | YES | N/A | N/A | YES | YES |
Our Open Documentation
|
|
|
|
|
|---|
The Team and Acknowledgements
Undergrads
|
Advisors
|
Acknowledgements
|
Funding
|
|---|