Ljubljana, Slovenia 2006/Project & Model
From 2006.igem.org
| Line 32: | Line 32: | ||
<h1>Selected project</h1> | <h1>Selected project</h1> | ||
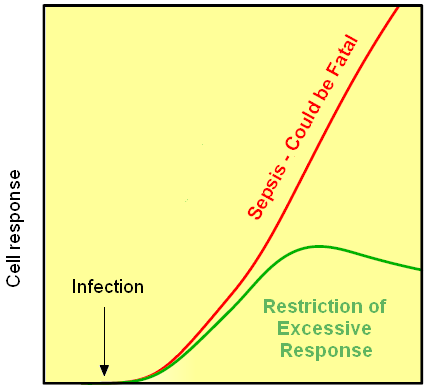
| - | [[Image:sepsa.png|250px|thumb|Figure | + | [[Image:sepsa.png|250px|thumb|Figure 4: Infection can lead to sepsis, which is in many cases fatal, but the negative feedback would decrease the imune response.]] |
<p>The basic idea of our project was to introduce a feedback loop in TLR signaling pathway, which would decrease the overwhelming response to the persistent or repeated stimulus with [[Ljubljana, Slovenia 2006/Terms & References#Terms|PAMP]]. However, completly shutting down the response to bacterial stimulation is not recommendable from the view of a host battling the infection. Ideally the feedback loop should decrease the response when it is too high but recover the responsiveness of the system after some time.</p> | <p>The basic idea of our project was to introduce a feedback loop in TLR signaling pathway, which would decrease the overwhelming response to the persistent or repeated stimulus with [[Ljubljana, Slovenia 2006/Terms & References#Terms|PAMP]]. However, completly shutting down the response to bacterial stimulation is not recommendable from the view of a host battling the infection. Ideally the feedback loop should decrease the response when it is too high but recover the responsiveness of the system after some time.</p> | ||
| Line 43: | Line 43: | ||
<h3>[[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] is the hub of the TLR signalling pathway</h3> | <h3>[[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] is the hub of the TLR signalling pathway</h3> | ||
<p> | <p> | ||
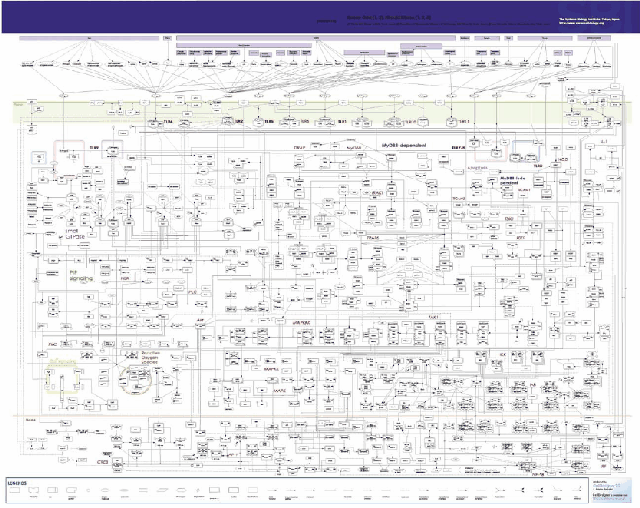
| - | Figure | + | Figure 5 represents the complexity of cell signalling mediated by TLRs. which contains more than 700 molecules and complexes (Oda, Kitano, Molecular Systems Biology 2006).</p> |
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| - | [[Image:model1.gif|thumb|center|800px|Figure | + | [[Image:model1.gif|thumb|center|800px|Figure 5: Comprehensive scheme of the Toll-like receptor signaling of microbial infection.]] |
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| - | <p>[[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] adapter protein is involved in the signal transduction immediately after ligand-induced TLR oligomerization. This adapter protein is common to most TLRs before the signaling network branches into several phosphpryation cascades. We selected this molecule as the most appropriate target of TLR signaling network engineering (Figures | + | <p>[[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] adapter protein is involved in the signal transduction immediately after ligand-induced TLR oligomerization. This adapter protein is common to most TLRs before the signaling network branches into several phosphpryation cascades. We selected this molecule as the most appropriate target of TLR signaling network engineering (Figures 6 and 7).</p> |
| - | [[Image:model2.gif|thumb|left|650px|Figure | + | [[Image:model2.gif|thumb|left|650px|Figure 6: [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] adapter protein connects the response of all TLRs and represents the »bottleneck« of the system (shaded area).]] |
| - | [[Image:model3.gif|thumb|right|200px|Figure | + | [[Image:model3.gif|thumb|right|200px|Figure 7: Schematic representation of [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]] as the hub of TLR signaling shaped like a bow tie .]] |
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| Line 61: | Line 61: | ||
<h3>Construction of the model</h3> | <h3>Construction of the model</h3> | ||
<p> | <p> | ||
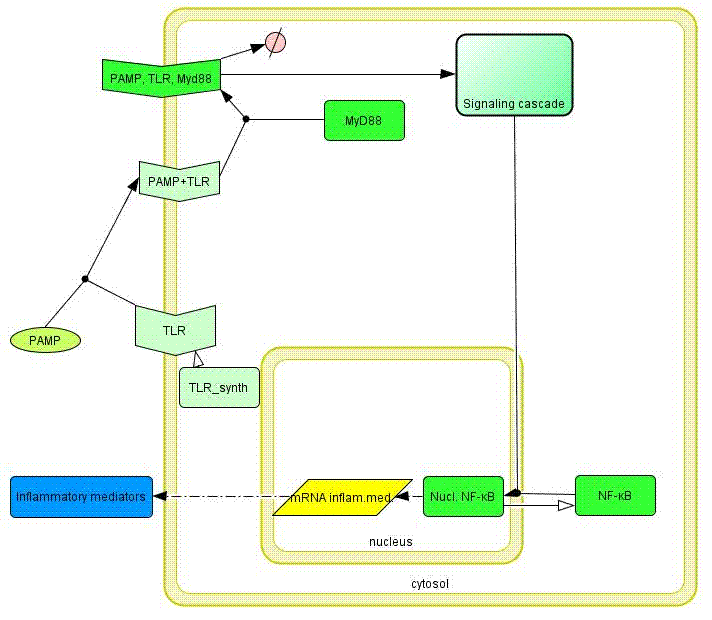
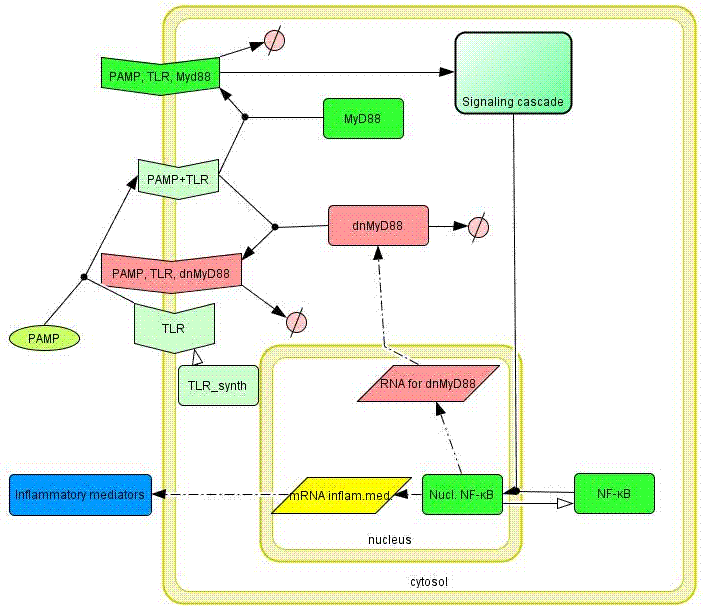
| - | Model of the TLR signaling was prepared in CellDesigner. Signaling steps, where the response remains the same were assembled into one block (Signaling cascade), whose activation directly affects the [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] translocation. Our engineered feedback loop is inserted as a transcriptional activation of [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] protein domain (red block in Figure | + | Model of the TLR signaling was prepared in CellDesigner. Signaling steps, where the response remains the same were assembled into one block (Signaling cascade), whose activation directly affects the [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] translocation. Our engineered feedback loop is inserted as a transcriptional activation of [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] protein domain (red block in Figure 8).</p> |
| - | [[Image:model12a.gif|left|thumb|400px|Figure | + | [[Image:model12a.gif|left|thumb|400px|Figure 8: Simplified model of Toll-like receptor signaling constructed in CellDesigner.]] |
| - | [[Image:model12.gif|right|thumb|400px|Figure | + | [[Image:model12.gif|right|thumb|400px|Figure 9: Model of TLR signaling with additional feedback loop for the inhibition of [[Ljubljana, Slovenia 2006/Terms & References#Terms|MyD88]].]] |
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| Line 72: | Line 72: | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
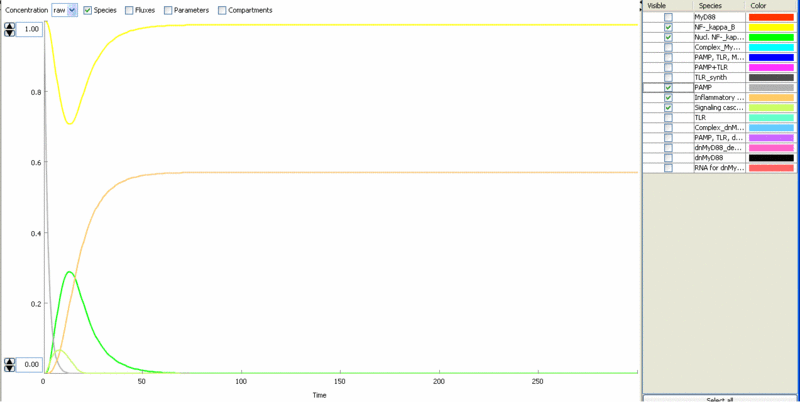
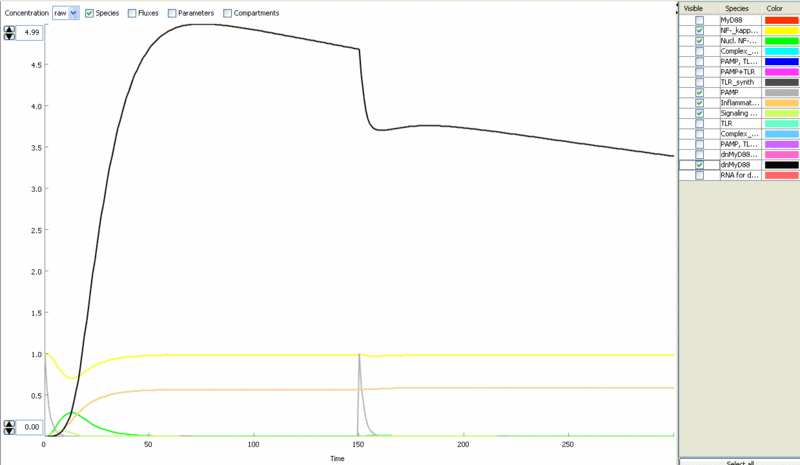
| - | [[Image:model6.gif|thumb|left|400px|Figure | + | [[Image:model6.gif|thumb|left|400px|Figure 10: Simulation of the cell response to the bacterial stimulus [[Ljubljana, Slovenia 2006/Terms & References#Terms|PAMP]] (grey) – transient translocation of the [[Ljubljana, Slovenia 2006/Terms & References#Terms|NF-κB]] into the nucleus (green), production of stimulatory mediators (orange).]] |
| - | [[Image:model7.gif|thumb|right|400px|Figure | + | [[Image:model7.gif|thumb|right|400px|Figure 11: Simulation of the repeated stimulation by [[Ljubljana, Slovenia 2006/Terms & References#Terms|PAMP]] (grey) - Amount of inflammatory mediators increases (orange).]] |
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| Line 79: | Line 79: | ||
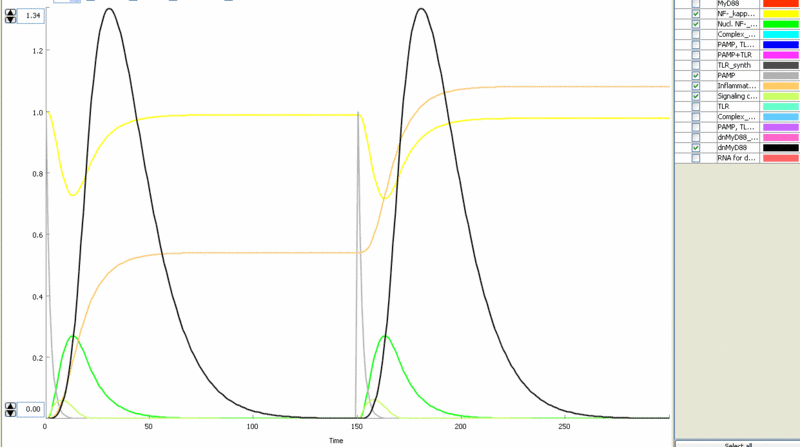
<p>Simulation of the TLR signaling response in the system with a feedback device results in the production of inhibitor ([[Ljubljana, Slovenia 2006/Terms & References|dnMyD88]], black) which inhibits cellular activation by a repeated stimulation. Cellular responsiveness resets to the normal response if the inhibitor is rapidly degraded, such as by the addition of a [[Ljubljana, Slovenia 2006/Terms & References|PEST]] motif.</p> | <p>Simulation of the TLR signaling response in the system with a feedback device results in the production of inhibitor ([[Ljubljana, Slovenia 2006/Terms & References|dnMyD88]], black) which inhibits cellular activation by a repeated stimulation. Cellular responsiveness resets to the normal response if the inhibitor is rapidly degraded, such as by the addition of a [[Ljubljana, Slovenia 2006/Terms & References|PEST]] motif.</p> | ||
| - | [[Image:model8.gif|thumb|left|400px|Figure | + | [[Image:model8.gif|thumb|left|400px|Figure 12: Addition of a feedback loop to start production of inhibitory [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]]( (black) – cell does not respond to the repeated stimulation.]] |
| - | [[Image:model9.gif|thumb|right|400px|Figure | + | [[Image:model9.gif|thumb|right|400px|Figure 13: Feedback loop with rapid degradation of the inhibitor ([[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] with PEST sequence - black) – responsiveness of the system is again completely restored.]] |
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
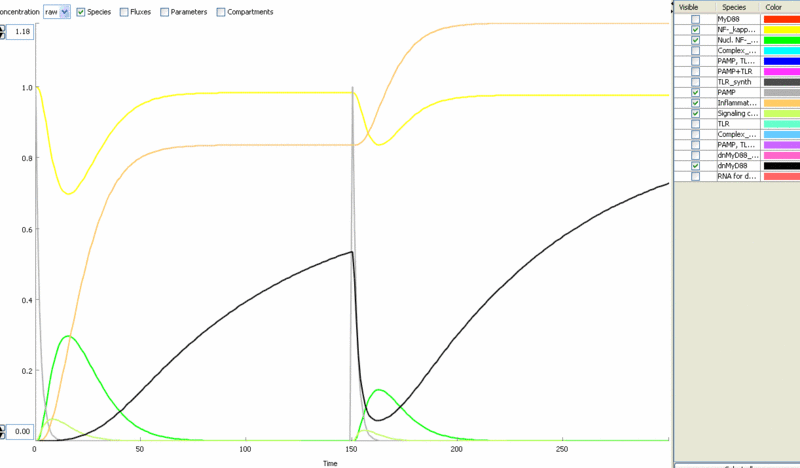
| - | [[Image:model10.gif|thumb|left|400px|Figure | + | [[Image:model10.gif|thumb|left|400px|Figure 14: Speed of the degradation of [[Ljubljana, Slovenia 2006/Terms & References#Terms|dnMyD88]] could modulate the responsiveness of the system to repeated (or continuous) challenge by TLR agonists.]] |
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
Revision as of 11:57, 30 October 2006
| Home | Background and Signalling Pathway | Methods | Results & Conclusions | Terms & References | Team members |
|---|
Contents |
Project ideas
Reasearch in the Laboratory of Biotechnology at the National institute of Chemisty, where most of our instructors come from and wheer most of the experimental work was conducted, is in the area of molecular immunology, specifically interactions in the Toll-like receptor signaling to microbial infections. This determined the selection of the eukaryotic system as our arena of synthetic biology "sandbox". besides, several contributions at the Synthetic biology meetings demonstrated that mammalian cell engineering
In the beginning we have discussed following project ideas:
- Inserton of a logical device into the signaling pathway to limit the cellular activation by bacterial infection - use of dominant negative versions of proteins involved in signaling pathway, which will arrest the signaling pathway thus preventing NF-κB (transcription factor mentioned above)to translocate to the nucleus - addition of degradation tags to this dominant negative proteins (PEST sequence), which would cause inhibition (negative feedback loop) to be temporal.
- Engineering of the response to pathogens/their PAMPs, which are otherwise not recognized by cells - for example a response to beta glucans of fungi.
- Engineering of novel interconnections between signaling pathways.
Selected project
The basic idea of our project was to introduce a feedback loop in TLR signaling pathway, which would decrease the overwhelming response to the persistent or repeated stimulus with PAMP. However, completly shutting down the response to bacterial stimulation is not recommendable from the view of a host battling the infection. Ideally the feedback loop should decrease the response when it is too high but recover the responsiveness of the system after some time.
Inhibition of the overwhelming response could be achieved if PAMP is simultaneously able to activate the immune response and the expression of a dominant-negative adapter protein, that would inactivate PAMP`s signaling pathway. Decreasing the lifetime of the dominant-negative inhibitior by the addition of a rapid degradation tag (PEST sequence) should inactivate the inhibition and restore the responsiveness of the immune system.
This idea is similar to the natural mechanism of tolerance, which is already present in living cells and which decreases the response to repeated bacterial stimulations. This natural tolerance is activated slowly, on the order of days and operates through several different mechanisms (Figure). Our feedback mechanism (i.e. artificial tolerance) should decrease the response within hours and thus "attack" the signaling pathway at the point, which has not been used in the natural system.
Model of the TLR signalling triggered by bacterial infection
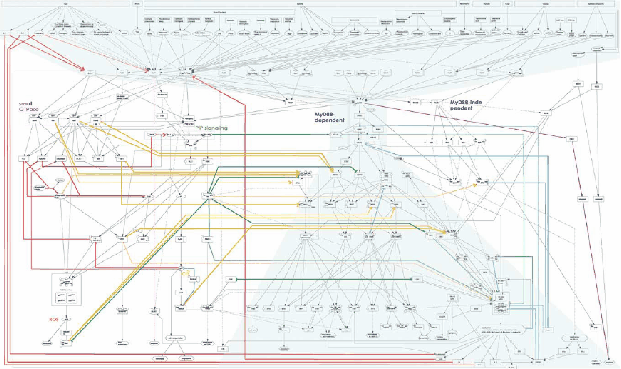
MyD88 is the hub of the TLR signalling pathway
Figure 5 represents the complexity of cell signalling mediated by TLRs. which contains more than 700 molecules and complexes (Oda, Kitano, Molecular Systems Biology 2006).
MyD88 adapter protein is involved in the signal transduction immediately after ligand-induced TLR oligomerization. This adapter protein is common to most TLRs before the signaling network branches into several phosphpryation cascades. We selected this molecule as the most appropriate target of TLR signaling network engineering (Figures 6 and 7).
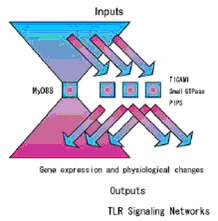
Construction of the model
Model of the TLR signaling was prepared in CellDesigner. Signaling steps, where the response remains the same were assembled into one block (Signaling cascade), whose activation directly affects the NF-κB translocation. Our engineered feedback loop is inserted as a transcriptional activation of dnMyD88 protein domain (red block in Figure 8).
Simulation of cellular response to TLR stimulation by a pulse of PAMP (e.g. flagellin). Signaling cascade results in the transient translocation of NF-κB into the cell nucleus (green) and in the production of inflammatory mediators. Repeated pulse after some time results in the repeated activation and accumulation of inflammatory mediators.
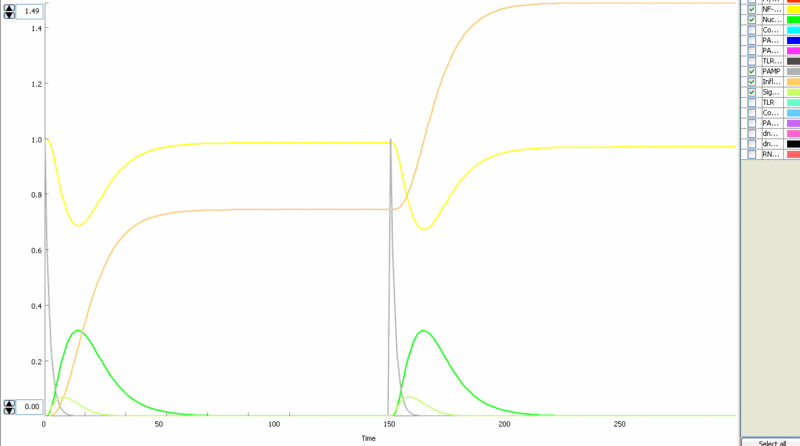
Simulation of the TLR signaling response in the system with a feedback device results in the production of inhibitor (dnMyD88, black) which inhibits cellular activation by a repeated stimulation. Cellular responsiveness resets to the normal response if the inhibitor is rapidly degraded, such as by the addition of a PEST motif.
| Home | Background and Signalling Pathway | Methods | Results & Conclusions | Terms & References | Team members |
|---|