McGill Repressilator M and M
From 2006.igem.org
| Line 3: | Line 3: | ||
<center>[[Image:Banner.gif]]</center> | <center>[[Image:Banner.gif]]</center> | ||
| - | <center>[[Image:Repressilator_plasmids. | + | <center>[[Image:Repressilator_plasmids.gif]][/center] |
<b>Methods and Materials</b> | <b>Methods and Materials</b> | ||
Revision as of 04:31, 28 October 2006
  [/center] [/center]
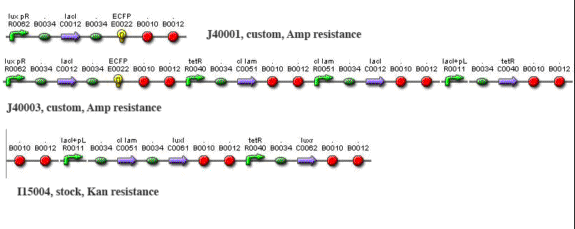
Methods and Materials Cloning Cloning by restriction enzyme digestion followed standard procedures. To create J40001, we ligated S0100 into the E0422 plasmid by cutting S0100 with EcoRI and SpeI and cutting E0422 with EcoRI and Xba1. We then ligated S0100/E0422 into the R0062 plasmid by cutting S0100/E0422 with XbaI and PstI, and by cutting R0062 with SpeI and PstI. To create J40002, we cut out the entire brick of I5610 with EcoRI and SpeI and ligated it to J40001 cut with EcoRI and XbaI. Ligations were performed using T4 DNA ligase at room temperature for 1-3 hours. Transfection and induction For Test system #1, MJF Lac- cells were transformed with I15004 and J40001. For test system #2, cells were transformed with I15004 and J40002. In all cases cells were seeded overnight in minimal medium containing ampicillin and kanamycin (minimal medium composition w/v: 0.2% ammonium sulfate,1.5% dipotassium phosphate, 0.6% monopotassium phosphate, 0.1% sodium citrate, 0.02% magnesium phosphate; supplemented with 5% dextrose and 0.01% yeast extract). They were then diluted 20 fold and grown to O.D. 0.3 at 600 nm. At this stage they were synchronized with 1 mM IPTG and incubated for another 1.5 h. Cells were then washed and resuspended in a smaller volume to increase cell density and interaction. Imaging Imaging was performed on a Zeiss510 confocal microscope with a heated stage. Bacteria were suspended in a 1-2% solution of low-melting-point (LMP) agarose in minimal medium, melted in the microwave and cooled to < 50 C. Before hardening, the solution was pipetted into the bottom of an imaging-quality glass bottom tissue 35-mm culture dish (MatTek) and allowed to solidify. 1-2 mL of minimal medium were then applied over the agarose plug to maintain nutrients and temperature over the imaging period. The dish was placed into a heated stage (Warner Instruments) and maintained at 32 C. The 458 nm line of an Ar laser was used to excite CFP and YFP; the output frequencies monitored were 480-520 (green channel) and 540-600 (yellow channel). The objective lens was a Nikon PlanFluor 63x oil. Imaging was performed every 2-5 minutes for 8-10 hours. Data were analyzed using ImageJ. Simulations Simulations were performed using BioNetS for MacOSX coupled to DataTank.
|