Missouri Western State University 2006
From 2006.igem.org
(→'''hix BioBricks''') |
(→'''hix BioBricks''') |
||
| Line 162: | Line 162: | ||
CCCCCTGCAGGCGGCCGCTACTAGTTTATCAAAAACCATGGTTTTTGATAATCTAGAAGCGGCCGCGAATTCCCCC | CCCCCTGCAGGCGGCCGCTACTAGTTTATCAAAAACCATGGTTTTTGATAATCTAGAAGCGGCCGCGAATTCCCCC | ||
| + | |||
| + | '''Backwards Parts''' | ||
| + | |||
| + | Because we wish to start with elements that are reversed (upside down pancakes), we need to think about how to get backwards parts. That is, parts that have the Prefix, then reversed element (promoter, term, CDS, RBS), then Suffix. Several possible ways: | ||
| + | |||
| + | # Some parts are backwards in the registry - terminators, for eg. | ||
| + | # Parts that are not long could be synthesized backwards. | ||
| + | # If one pancake can be flipped, reversed parts can be made. This may actually be a good contribution, since it could be applied to reverse any part between two hix sites. | ||
| + | |||
| + | For example, the RBS Bba_B0034 has this sequence: | ||
| + | |||
| + | gaattcgcggccgcttctaga g'''aaagaggagaaa'''t actagtagcggccgctgcag | ||
| + | |||
| + | It could be redesigned with the RBS reversed: | ||
| + | |||
| + | gaattcgcggccgcttctaga g'''tttctcctcttt'''t actagtagcggccgctgcag | ||
| + | |||
'''Process of Creating Synthetic Pancake:''' | '''Process of Creating Synthetic Pancake:''' | ||
Revision as of 22:12, 1 June 2006
Front Row (left to right) Adam Brown, Trevor Butner, Brad Ogden, Marian Broderick, Todd Eckdahl.
Back Row (left to right) Jeff Poet, Eric Jessen, Kelly Malloy
Contents |
Students
• Marian Broderick [1]
• Adam Brown [2]
• Trevor Butner [3]
• Lane Heard [4]
• Eric Jessen [5]
• Kelly Malloy [6]
• Brad Ogden [7]
• Kayla Ragsdale [8]
Faculty
• Todd Eckdahl [http://staff.missouriwestern.edu/~eckdahl/], Department of Biology, [9]
• Jeff Poet [http://staff.missouriwestern.edu/~poet/], Department of Computer Science, Mathematics, and Physics, [10]
Shipping Address: Todd Eckdahl, Biology Department, Missouri Western State University, 4525 Downs Drive, Saint Joseph, MO, 64507
Research Links
Goodman et al. 1973-1979. The Pancake Problems [http://www.math.uiuc.edu/~west/openp/pancake.html]
Nanassy and Hughes. 1998. In Vivo Identification of Intermediate Stages of the DNA Inversion Reaction Catlyzed by the Salmonella Hin Recombinase [http://www.genetics.org/cgi/content/abstract/149/4/1649]
Lim et al. 1997. Hin-mediated Inversion on Positively Supercoiled DNA [http://www.jbc.org/cgi/content/full/272/29/18434]
Hughes et al. 1992. Sequence-specific interaction of the Salmonella Hin recombinase in both major and minor grooves of DNA [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=1628628]
Nanassy and Hughes. 2001. Hin Recombinase Mutants Functionally Disrupted in Interactions with Fis [http://jb.asm.org/cgi/content/full/183/1/28?view=long&pmid=11114897]
Lee et al. 1998. In Vivo Assay of Protein-Protein Interactions in Hin-Mediated DNA Inversion [http://jb.asm.org/cgi/content/abstract/180/22/5954]
Merickel et al. 1998. Communication between Hin recombinase and Fis regulatory subunits during coordinate activation of Hin-catalyzed site-specific DNA inversion [http://www.genesdev.org/cgi/content/abstract/12/17/2803]
Haykinson and Johnson. 1993. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=8508775]
Vasquez et al. 2004. Tangle Analysis of Gin Site-specific Recombination [http://math.berkeley.edu/~mariel/Vazquez_Gin04.pdf]
Merickel and Johnson. 2004. Topological analysis of Hin-catalysed DNA recombination in vivo and in vitro [http://www.blackwell-synergy.com/doi/abs/10.1046/j.1365-2958.2003.03890.x]
R.C. Johnson and M.F. Bruist. 1989. Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction [http://www.pubmedcentral.gov/picrender.fcgi?tool=pmcentrez&blobtype=pdf&artid=400990]
Lim and Simon. 1992. The Role of Negative Supercoiling in Hin-mediated Site-Specific Recombination[http://www.jbc.org/cgi/content/abstract/267/16/11176]
The Pancake Tossing World Record[http://www.recordholders.org/en/records/pancake.html]
Cool site for Breakfast [http://www.cut-the-knot.org/SimpleGames/Flipper.shtml]
iGEM iDeas
Powerpoint with two possible project ideas. [http://staff.missouriwestern.edu/~eckdahl/Research/Synthetic_Biology_2006/IGEMideas%2005-29-06.ppt]
Parts to Make:
HixL: TTCTGGAAAA CCAAGGTTTT TGATAA
HixR: TTATCAAAAA CCTTCCAAAA GGAAAA
HixC: TTATCAAAAA CCATGGTTTT TGATAA
HixC1: TTCTGGAAAACC AT CCAAAAGGAAAA
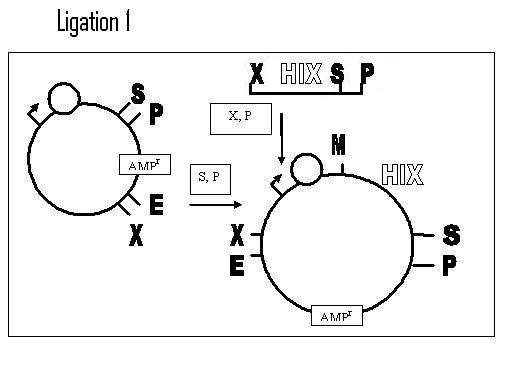
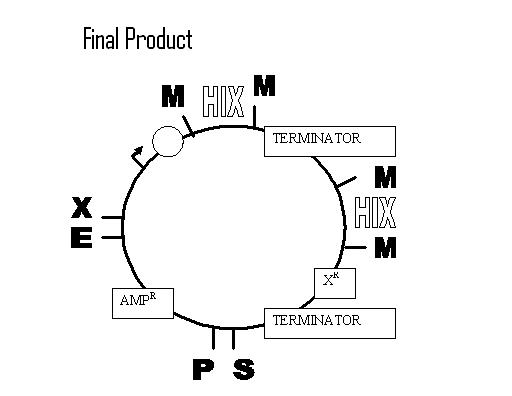
Steps
1. Design/Order DNA – Hix L, R, & C
- Want to do a Left to Right BioB addition: Promoter + RBS w/AmpR -->Hix site-->
Term-->Hix(C)site-->TetR + Term
2. On receiving – restrict/ligate BioBricks into plasmid w/resistance gene on one side
a. Promoter, RBS BioB
b. TetR BioB (Backwards)
c. Term BioB
3. Transform E. coli
E = G/AATC
X = T/CTAGA
P = CGAT/CG
S = A/CTAGT
hix BioBricks
Hey, it's your Davidson pals.We noticed that your biobricks do not have the Not1 site between E and X in the prefix and S and P in the suffix Thanks.
E - N - X - hix - S - N - P
Does there have to be a T between NotI and XbaI and an A between SpeI and NotI? All the parts we looked at (10 of them) had this.
HixL
GGGG GAATTC GCGGCCGC T TCTAGA TTCTTGAAAACCAAGGTTTTTGATAA ACTAGT A GCGGCCGC CTGCAG GGGG
HixR
GGGG GAATTC GCGGCCGC T TCTAGA TTATCAAAAACCTTCCAAAAGGAAAA ACTAGT A GCGGCCGC CTGCAG GGGG
HixC
GGGG GAATTC GCGGCCGC T TCTAGA TTATCAAAAACCATGGTTTTTGATAA ACTAGT A GCGGCCGC CTGCAG GGGG
Oligos to order
HixL_top
GGGGGAATTCGCGGCCGCTTCTAGATTCTTGAAAACCAAGGTTTTTGATAAACTAGTAGCGGCCGCCTGCAGGGGG
HixL_bottom
CCCCCTGCAGGCGGCCGCTACTAGTTTATCAAAAACCTTGGTTTTCAAGAATCTAGAAGCGGCCGCGAATTCCCCC
HixR_top
GGGGGAATTCGCGGCCGCTTCTAGATTATCAAAAACCTTCCAAAAGGAAAAACTAGTAGCGGCCGCCTGCAGGGGG
HixR_bottom
CCCCCTGCAGGCGGCCGCTACTAGTTTTTCCTTTTGGAAGGTTTTTGATAATCTAGAAGCGGCCGCGAATTCCCCC
HixC_top
GGGGGAATTCGCGGCCGCTTCTAGATTATCAAAAACCATGGTTTTTGATAAACTAGTAGCGGCCGCCTGCAGGGGG
HixC_bottom
CCCCCTGCAGGCGGCCGCTACTAGTTTATCAAAAACCATGGTTTTTGATAATCTAGAAGCGGCCGCGAATTCCCCC
Backwards Parts
Because we wish to start with elements that are reversed (upside down pancakes), we need to think about how to get backwards parts. That is, parts that have the Prefix, then reversed element (promoter, term, CDS, RBS), then Suffix. Several possible ways:
- Some parts are backwards in the registry - terminators, for eg.
- Parts that are not long could be synthesized backwards.
- If one pancake can be flipped, reversed parts can be made. This may actually be a good contribution, since it could be applied to reverse any part between two hix sites.
For example, the RBS Bba_B0034 has this sequence:
gaattcgcggccgcttctaga gaaagaggagaaat actagtagcggccgctgcag
It could be redesigned with the RBS reversed:
gaattcgcggccgcttctaga gtttctcctctttt actagtagcggccgctgcag
Process of Creating Synthetic Pancake: