Proposal & Approach
From 2006.igem.org

| Home | Backround and Signalling Pathway | Anticipated Results & Significance | Troubleshooting, References & Sponsors | Team members |
|---|
Contents |
Project proposal
In the beginning we have discussed following project ideas:
- New mechanism of tolerance - use of inhibitors that interfere with NF-kappaB (transcription factor mentioned above) - inhibition with dominant negative proteins involved in signaling pathway, this proteins could be labeled with degradation tags (PEST sequence) and inhibition would be temporal (negative feedback loop)
- Cell response to pathogen, that cells usualy can not recognise - for example response to beta glucans of fungi
- Find a connection/shortening of signaling pathways to make it more efficient and include more responses
Selected project proposal
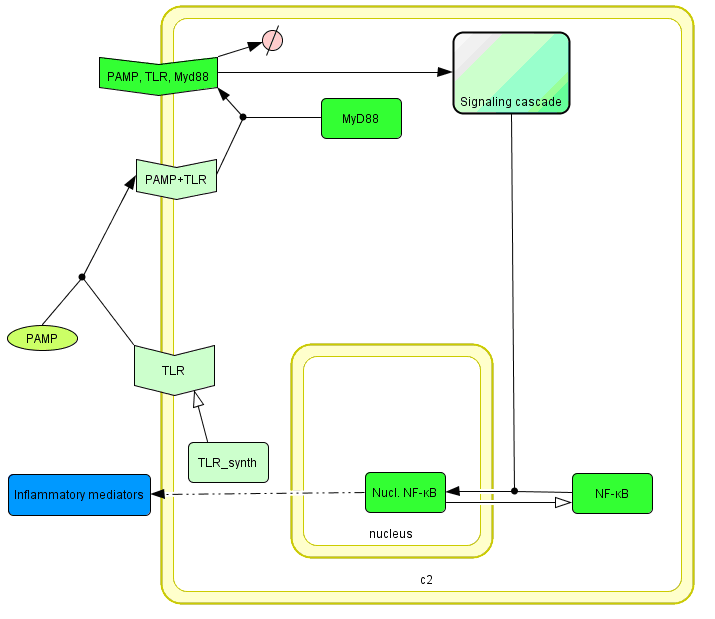
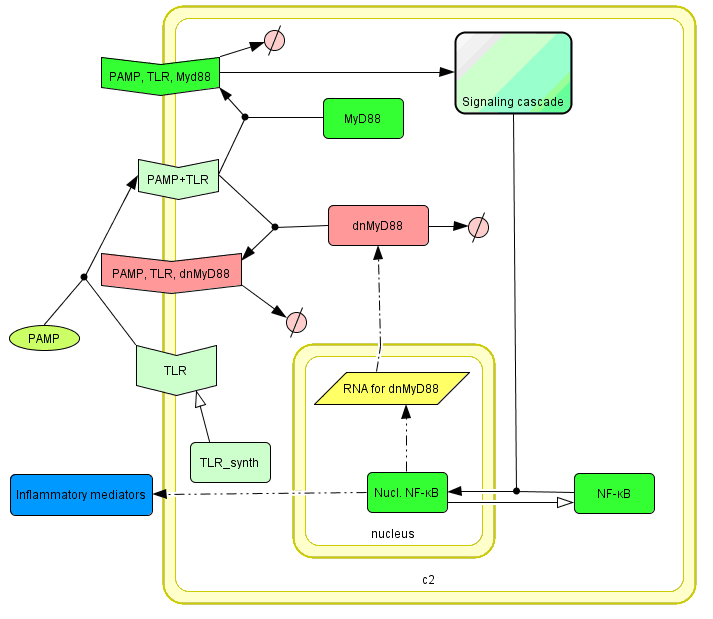
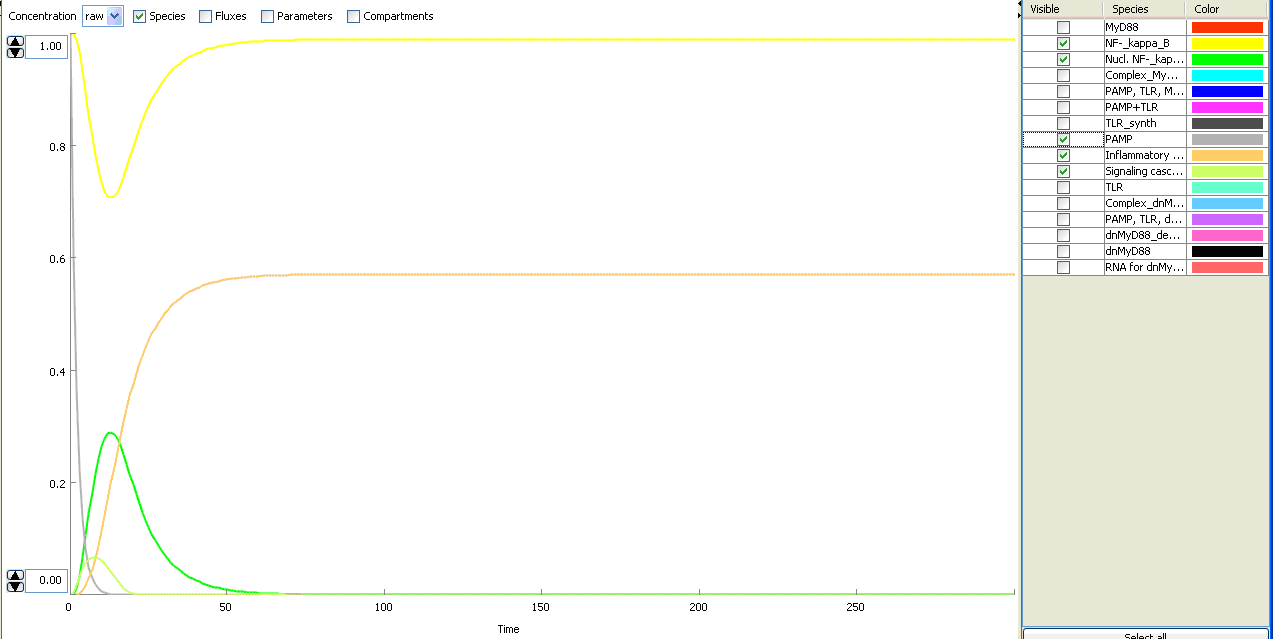
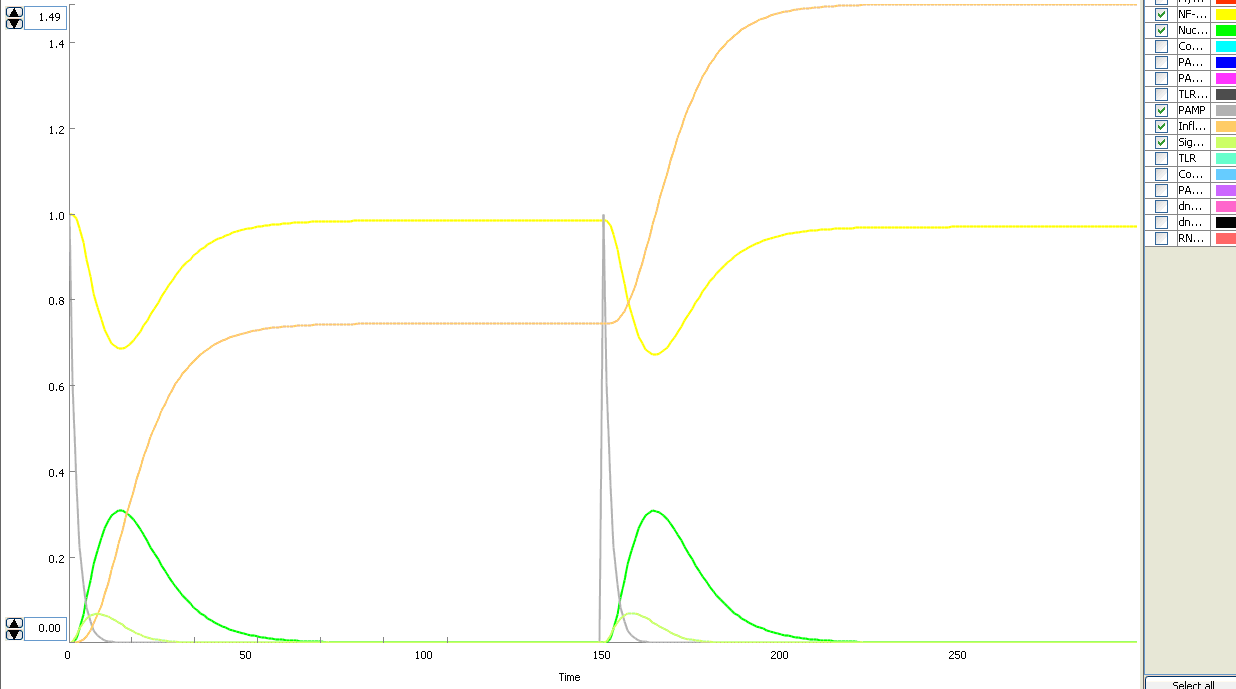
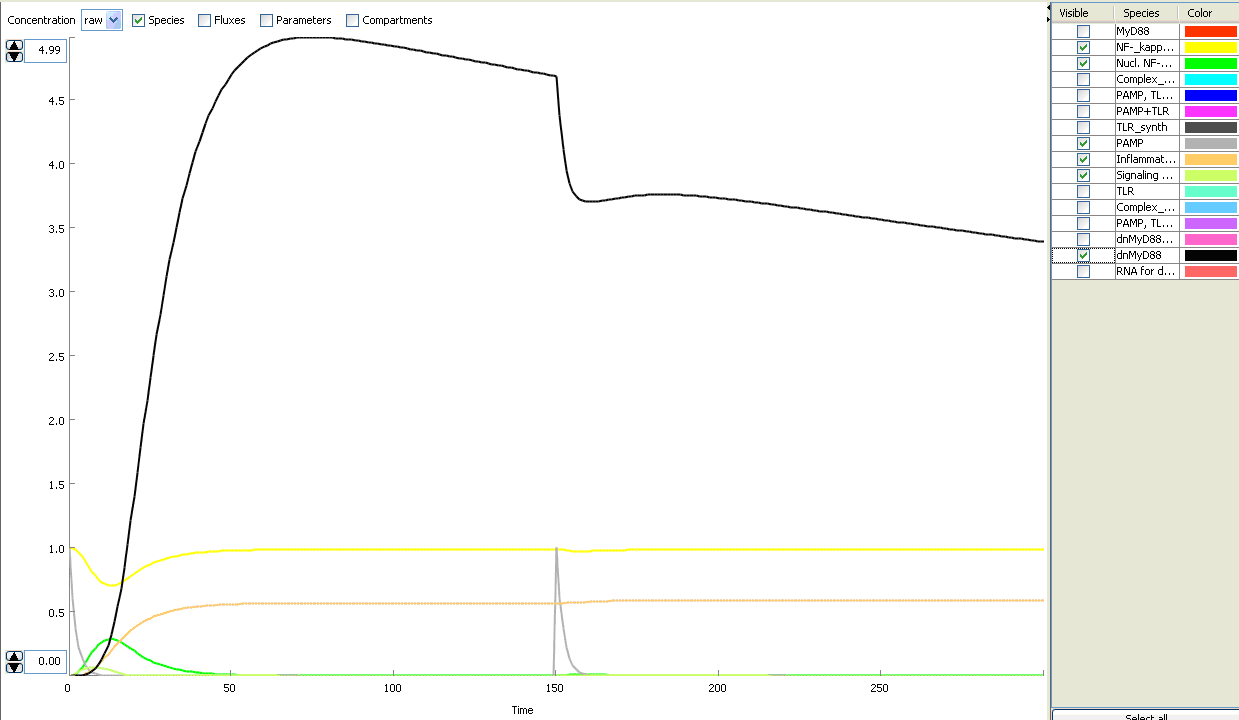
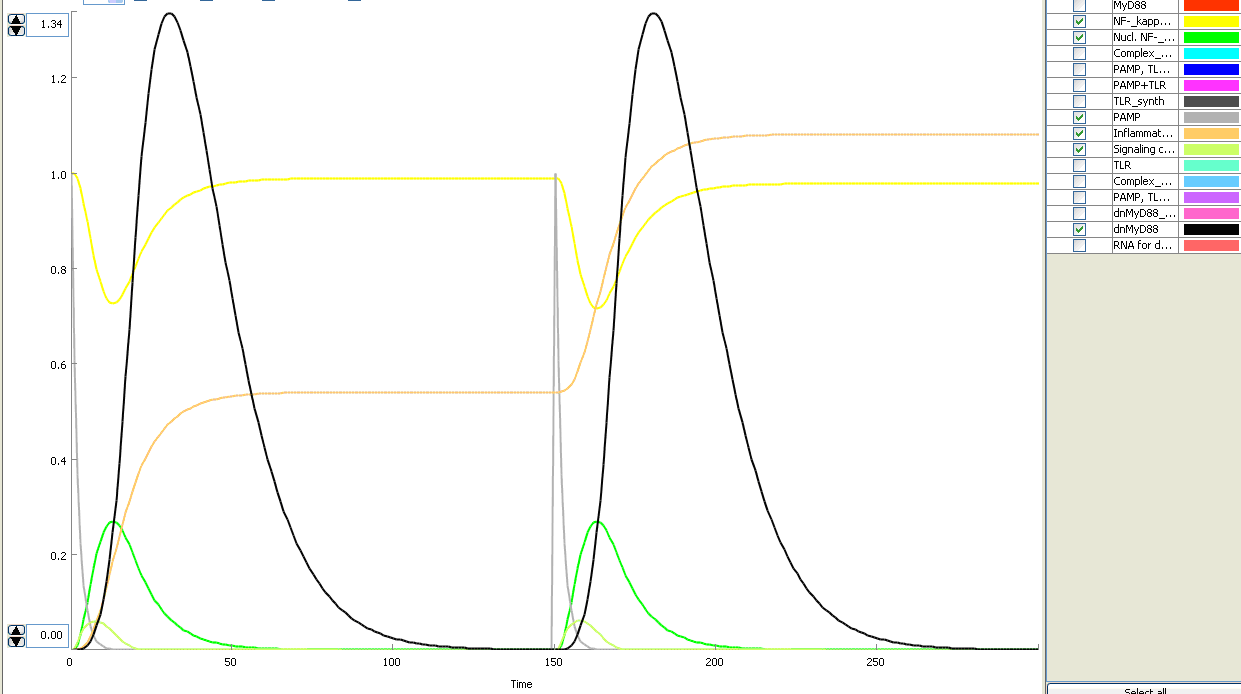
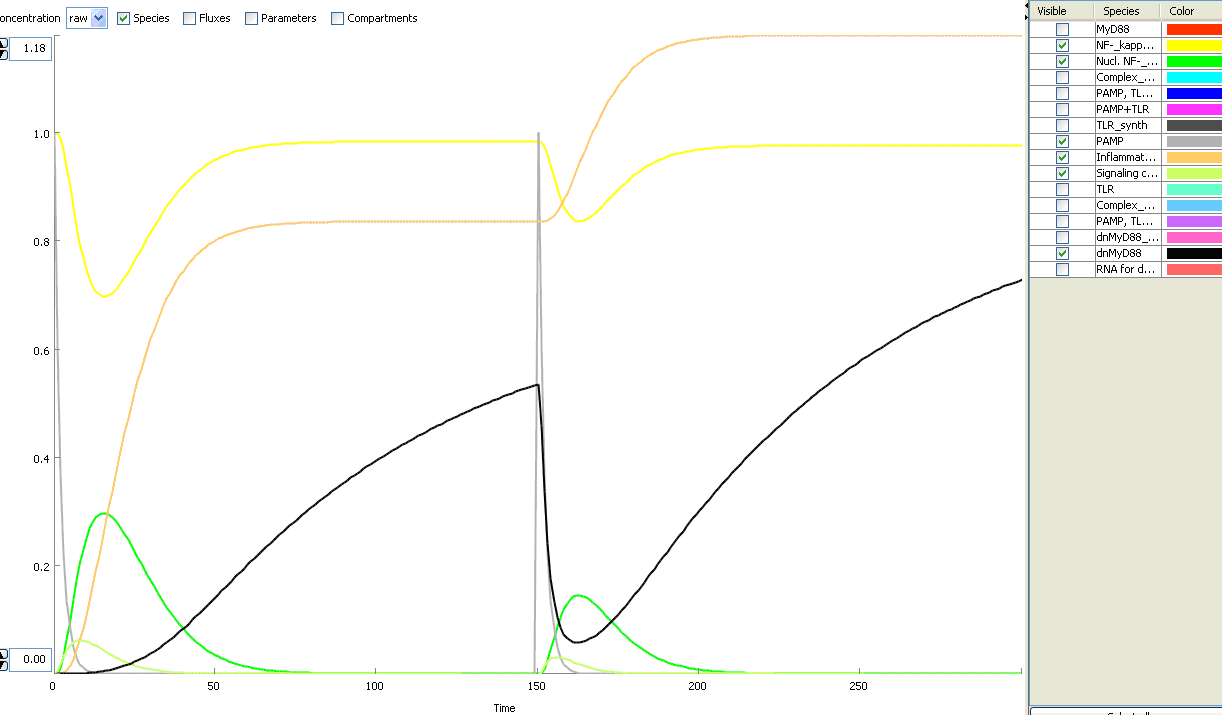
The basic idea of our project was to introduce the feedback loop,which would decrease the response to the persistent or repeated stimulus. However completly shutting down the response at bacterial stimulation is not a good solution. Ideally the feedback loop should decrease the response when it is too high but recover the responsiveness of the system after some time. Inhibition of the response could be achieved by activating the expression of the dominant-negative adapter protein, that inactivates the signaling pathway. Decreasing the lifetime of the dominant-negative inhibitior by the addition of rapid degradation tag (PEST sequence) should inactivate the inhibition and reset (restore) the normal responsiveness of the immune system.
This idea is similar to the natural mechanism of tolerance, which is already present in living cells and which decrease the response to repeated bacterial stimulation. This natural tolerance is activated slowly, on the order of days and operates through several different mechanisms (Figure). Our feedback mechanism (i.e. artificial tolerance) should decrease the response within hours and thus "attack" the signaling pathway at the point, which has not been used in the natural system.
Approach
Mathematical model of signaling
Parts design
At first we had to design primers to replicate a desired DNA fragment. In primers we included restriction sites - on left site XbaI and on the right site SpeI, NcoI and PstI. We cloned that part in to BioBrick plasmids with ccdB domain to get all restriction sites needed for BioBrick assembly. We had to design all parts de novo, since no parts like promoters, terminators, desired proteins for signaling pathway modification, degradation flags and reporters had been designed so far - neither to work in mammalian cells. List of desired constructs is shown below. For our use we designed a special vector (BBa_J52017) with terminator to simplify constructs assembly. All our composite parts (promoter plus part) were then cloned in this vector. We also needed fusion proteins e.g. dnMyd88-rLuc-PEST (BBa_J52013) - that is our dominant negative protein linked with reporter and degradation flag. This parts are designed like basic parts - not composite, although they are fusion proteins. Between proteins there is a 6 aminoacid long linker because only 2 aminoacid long linker formed during biobrick assembly could affect protein folding. We introduced a six amino acids long linker in between protein - reporter and reporter - degradation flag with primers using PCR Overlap Extension method. These parts were then combined with promoter (NF-kappaB) in BioBrick assembly technique. The part was then inserted in vector with terminator and ready to use in human cells (HEK 293).
| Registration number | Part's Name | Vector |
|---|---|---|
| BBa_J52008 | BBa_J52012 | rluc | pSB1AK3 | </tr>
| BBa_J52011 | ||
| BBa_J52013 | rluc-link-pest191 | pSB1AK3 |
| BBa_J52023 | MyD88-link-rluc-verija2 | pSB1AK3 |
| BBa_J52010 | MyD88-link-rluc-link-PEST191 | pSB1AK3 |
| BBa_J52014 | NFkB+rluc-link-PEST191 | pSB1AK3+TER |
| BBa_J52018 | NFkB | pSB1AC3 |
| BBa_J52036 | NFkB+MyD-link-Rluc | pSB1AK3+TER |
| BBa_J52024 | NFkB+rluc | pSB1AK3+TER |
| BBa_J52034 | NFkB+MyD | pSB1AK3+TER |
| BBa_J52038 | NFkB+MyD-Rluc-PEST191 | pSB1AK3+TER |
| BBa_J52039 | CMV | pSB1A2 |
| BBa_J52642 | CMV+rluc | pSB1AK3+TER |
| BBa_J52648 | CMV+rluc-PEST191 | pSB1AK3+TER |
| BBa_J52040 | GFP | pSB1AK3 |
| BBa_J52035 | CMV+GFP | pSB1AK3+TER |
| BBa_J52026 | CMV+GFP-PEST191 | pSB1AK3+TER |
| BBa_J52028 | MyD | pSB1AK3 |
| BBa_J52029 | MyD-GFP | pSB1AK3 |
| BBa_J52027 | GFP-PEST191 | pSB1AK3 |
| BBa_J52019 | NFkB+GFP-PEST191 | pSB1AK3+TER |
| BBa_J52021 | NFkB+MyD-GFP | pSB1AK3+TER |
| BBa_J52022 | TRAF6-GFP | pSB1AK3 |
| BBa_J52016 | NFkB+TRAF6-GFP | pSB1AK3+TER |
| BBa_J52017 | terminator | pSB1AK3 |
Transfection
In September and October we started to work on transfection of our constructs into human embrional kidney cells (HEK293). and three detection system mentioned below. At first we had to optimize the methods (read articles, test negative and positive controls) and learn how to work with human cells. Experiments are still in progress. Despite transfection with our construct, we also have to transfect cellc with TLRs, because strain HEK293 expresses only TLR3 and TLR6. Sepsis is usually response to pathogenic Gramm negative bacteria. Their outer membrane contains LPS (lipoproteins) that is recognized by TLR4 and accessory molecule MD2. At the beginning we transformed all cells with our constructs and additional plasmids, one coding TLR4 receptor and another MD2. Very soon we found out that our plasmids were contaminated with LPS of E. coli (strain DH5alpha used for transformation), since there was no difference in response between stimulated (with LPS) and unstimulated cells. To overcome this problem, now we are using TLR5 receptor. This receptor detects bacterial flagelin. Signal transfer through this receptor does not depend on presence of LPS.
Detection systems
We are testing our hypothesis using three different detection systems. All of them has different approach, however we are expecting the same result – synthesis of dominant negative protein (MyD88 or TRAF6), blocking of signalling pathway and consequently transcription termination of dominant negative protein. The construct (parts) we have made are designed for each detection system respectively.
Requests for the optimal detection system were:
- velocity;
- sensitivity;
- paralelization;
- optical signal;
- in vivo detection;
- low price.
On the basis of those requests we have decided for the following detection systems: flow cytometry, luminometry and ELISA. Members of those subgroups incharged for each system are :
Komentarji: Časi ... pišemo, kot da še delamo al v pretekliku. Jest sm pisala, kot da še, samo se mi ne zdi primerno oz. bi na konc (predn gremo) mogl spremenit.
Beseda signalling: kaj je bolj prav. V literaturi se pojavlja t enim ali dvema L-jem. Word smatra za pravilno besedo z enim L.
-popravi tabelo -označi, kaj pomeni kateri part -v tabeli vstavi linke za BioBricke
| Home | Backround and Signalling Pathway | Anticipated Results & Significance | Troubleshooting, References & Sponsors | Team members |
|---|