ETH Zurich 2006
From 2006.igem.org
Christophe (Talk | contribs) |
Christophe (Talk | contribs) |
||
| Line 85: | Line 85: | ||
= [[Introduction]] = | = [[Introduction]] = | ||
| - | + | The past few years have seen the emergence of the field of synthetic biology, in which functional units are designed and built into living cells to generate a particular behaviour, and ultimately to better understand Life's mechanisms. Previous efforts include the creation of gene circuits that generate oscillating behaviour ([[Elowitz00]]), toggle switch functionality ([[Atkinson03]]), artificial cell-cell communication ([[Bulter04]]) or pattern-forming behaviour ([[Basu2005]]). The present document describes the design and realization of a gene circuit that counts to 4. | |
= [[Modeling and overall design|Modeling]] = | = [[Modeling and overall design|Modeling]] = | ||
Revision as of 07:57, 21 October 2005
Short Overview. The project of the ETH Zurich team consists of the design and implementation in vivo of a gene circuit that can count up to 4. In essence, the counter uses two toggle switches, each storing 1 bit, to keep track of the 4 states. The design of the counter is highly modular, with the hope that it can be included as a unit in larger circuits, and also combined with further counter instances to keep track of a much larger number of states, up to (2^(n+1)) with n units. To facilitate further developments and integration to other projects, the counter is available in form of BioBricks. Among many exciting applications, the availability of a counter enables the execution of sequential instructions, and paves the way for the execution of artifical programs inside living cells.
Contents |
News
- 2005.10.18 The parts for the actual INPUT-module are ready, thanks to the hard work of Giorgia, Hervé, and Martje (not all test/debbuging parts though)
- 2005.10.07 Message from Blue Heron: Sequences for NOR are synthesized and will be verified and assembled next week.
- 2005.09.23 Sequences for NOR module ordered from Blue Heron
Organisation
People
Students
| Simon Barkow | Christophe Dessimoz | Zlatko Franjcic |
| Dominic Frutiger | Robin Künzler | Urs A. Müller |
| Jonas Nart | Kristian Nolde | Alexander Roth |
| Tamara Ulrich | Giorgia Valsesia | Herve Vanderschuren |
Supervisors
| Jörg Stelling | Sven Panke | Eckart Zitzler |
Advisors
| Uwe Sauer | Martin Fussenegger | Andreas Hierlemann |
| Kay-Uwe Kirstein | Ruedi Aebersold |
Timeline & Tasks
Groups
Modeling
- Members: Tamara, Kristian, Zlatko
This group is focused on mathematical modeling, illustration, simulations and overall design. There are no permanent members for this group. Anyone can join and leave the group anytime he/she wants. A group coordinator will be assigned at the first meeting.
- Meetings:
empty
Input Module
- Members: Christophe, Dominic (coordinator), Giorgia, Herve, Zlatko
- Meetings:
log 2005-08-17: Wednesday, 13:00 @ polyterrasse: Discussion of module, next steps, task distrib. log 2005-08-22: Monday, 15:00 @ polyterrasse: Discussion of biol. solutions. log 2005-09-12: Thursday, 17:30 teammeeting. ... multiple meetings, no log ... 2005-10-10: Monday, 10:00 @ intro to FACS system (Sven, Giorgia, Hervé, Dominic). 2005-10-11: Tuesday, 15:00 @ polyterrasse: planning test session (Giorgia, Dominic) log 2005-10-13: Thursday, 17:30 @ test session at Leica AOBS at LMC (Giorgia, Dominic) 2005-10-19: Wednesday, 14:00 @ planning of tests, giorgia, hervé, dominic)
NOR Module
- Members: Alex (coordinator), Jonas, Robin, Simon, Tamara, Urs
- Meetings:
log 2005.08.17, Wednesday, 15:30 2005.08.18, Thursday, 13:15 log 2005.08.22, Monday, 13:45 2005.08.25, Thursday 15:30 2005.08.29, Monday 14:00 2005.08.30, Tuesday 16:00 2005.09.06, Tuesday 08:00
Documentation: 'todo' list
Introduction
The past few years have seen the emergence of the field of synthetic biology, in which functional units are designed and built into living cells to generate a particular behaviour, and ultimately to better understand Life's mechanisms. Previous efforts include the creation of gene circuits that generate oscillating behaviour (Elowitz00), toggle switch functionality (Atkinson03), artificial cell-cell communication (Bulter04) or pattern-forming behaviour (Basu2005). The present document describes the design and realization of a gene circuit that counts to 4.
Modeling
Methods
Input-module
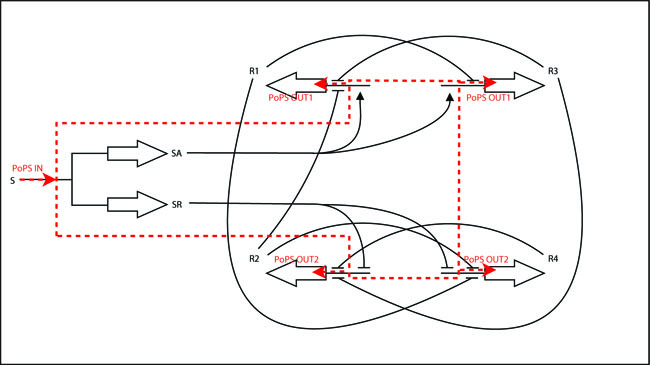
Module description: in a nutshell, this module has 2 system boundaries, both of which are characterized by PoPS. Its purpose is to take a single input PoPS and output 2 different PoPS. One of the outputs should be high and the other low when S is high and vice versa when S is low.
The iGEM-team at ETH decided to implement a modular and standardized Counter-system that can in principle be used to count any kind of biological event and generate any kind of output after a certain number N of events has been detected. The Counter-system can be split into 2 basic units, the NOR-module and the INPUT-module (outlined in red in the schematic below). This document deals with the implementation of the Input-module.
NOR Module
A NOR-module works as follows: There are two inputs and one output. The output is high if and only if the two inputs are low. In other words: The output is low whenever either one (or both) of the inputs are high.
Counter
Results
Discussion
Appendix
References
Modeling and illustration tools,
bulter04, atkinson03, bates05, keiler01, suetsugu03, sudesh00, römling02, ross91, sutherland01, Lai04, zogaj01, miller01, basu05, goryachev05, you04, Isalan01, Beerli02, Mani05, Beerli00, Beerli98, Dreier01, Dreier05, Klug05, Yang95, Segal99, Segal03,
Glossary
Previous Ideas
This is the brainstorming and previous ideas section. In this section you will find other projects that had been pursued, as well as random ideas without too much consideration of feasibility, etc.