EdinburghModeling
From 2006.igem.org
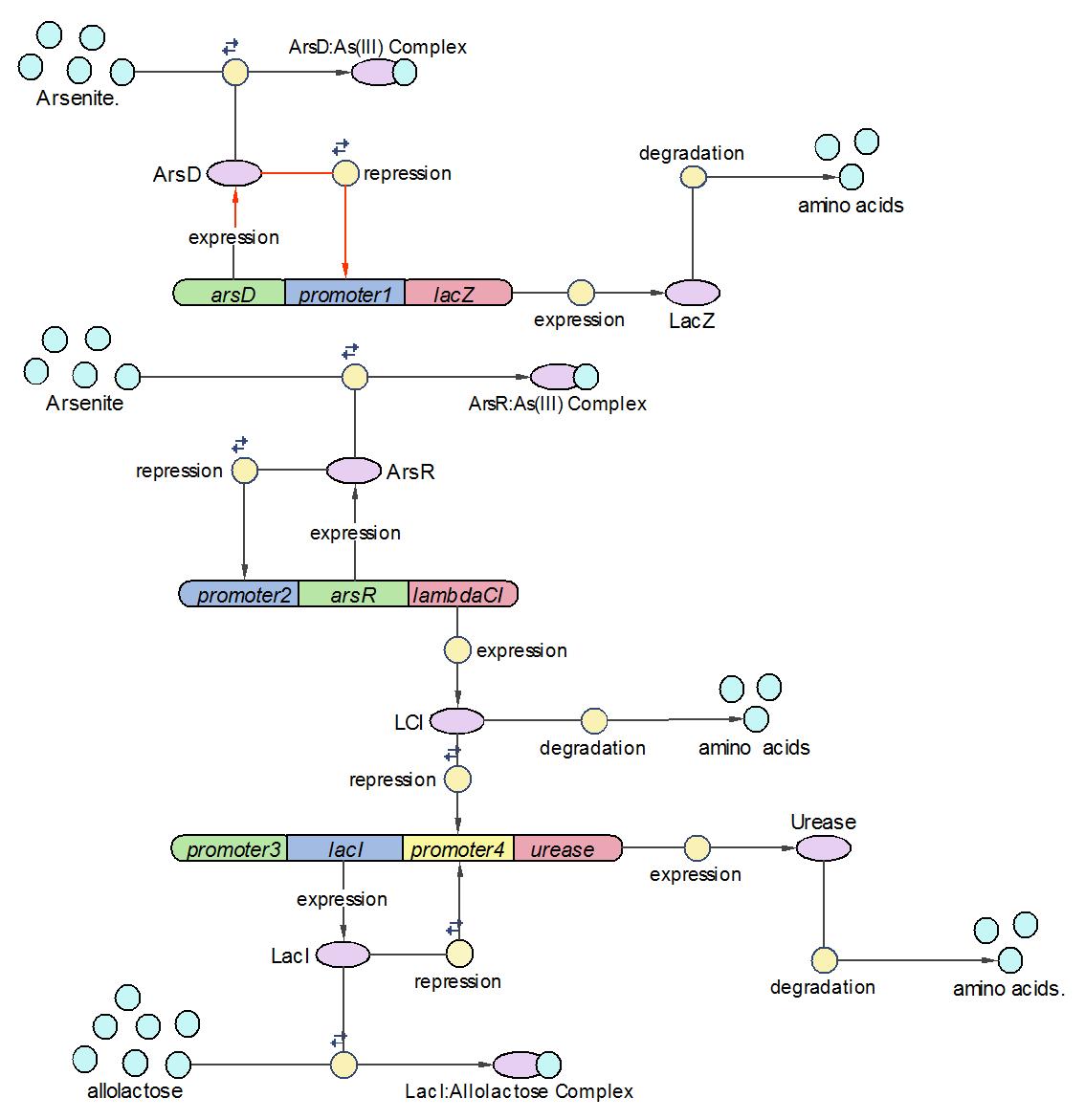
System Diagram of Arsenic Biosensor
Reaction set
| No. | Name | Equation | Rate Law | Parameters |
| 1 | ArsD binding to Arsenic (III) | ArsD+2As(III)=ArsD-2As(III) | Mass Action | K1=1000 K-1=0.65 |
| 2 | ArsD binding to promoter1 | 2 ArsD+promoter1=2ArsD-promoter1 | Mass Action | K2=10000, K-2=0.65 |
| 3 | ArsD degradation | ArsD->null | Mass Action | K3=0.05 |
| 4 | ArsD production | promoter1->promoter1+ArsD | Michaelis-Menton | V4m=0.5, K4m=75 |
| 5 | ArsR binding to Arsenic (III) | ArsR+2As(III)=ArsR-2As(III) | Mass Action | K5=1000, K-5=0.65 |
| 6 | ArsR binding to promoter2 | 2ArsR+promoter2=2ArsR-promoter2 | Mass Action | K6=10000, K-6=0.65 |
| 7 | ArsR degradation | ArsR->null | Mass Action | K7=0.05 |
| 8 | ArsR production | promoter2->promoter2+ArsR | Michaelis-Menton | V8m=10, K8m=25 |
| 9 | LacI binding to allolactose | LacI+allolactose=LacI-allolactose | Mass Action | K9=10000, K-9=0.1 |
| 10 | LacI binding to promoter4 | LacI+promoter4 = LacI-promoter4 | Mass Action | K10=1000, K-10=0.5 |
| 11 | LacI degradation | LacI->null | Mass Action | K11=0.1 |
| 12 | LacI production | promoter3->promoter3+LacI | Michaelis-Menton | V12m=0.5, K12m=40 |
| 13 | lacZ degradation | lacZ->null | Mass Action | K13=0.1 |
| 14 | lacZ production | promoter1->promoter1+lacZ | Michaelis-Menton | V14m=25, K14m=10 |
| 15 | LCI binding to promoter 4 | LCI+promoter4=LCI-promoter4 | Mass Action | K15=10000, K-15=0.5 |
| 16 | LCI degradation | LCI->null | Mass Action | K16=0.1 |
| 17 | LCI production | promoter2->promoter2+LCI | Michaelis-Menton | V17m=10, K17m=25 |
| 18 | Urease degradation | Urease->null | Mass Action | K18=0.1 |
| 19 | Urease production | promoter4->promoter4+Urease | Michaelis-Menton | V19m=10, K19m=40 |
Note: The units for first, second and third order rate constants are expressed in units of <math>second^{-1}</math>, nMol^{-1}second^{-1} and l^{2}nMol^{-2}second^{-1}respectively.