Davidson 2006
From 2006.igem.org
| Line 26: | Line 26: | ||
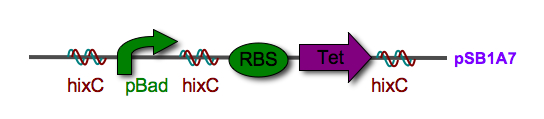
|[[Image:Jmol_Hin_tetrad_DNA.gif|thumb|250px|right|'''Figure 2''' 3-D structure of a Hin protein complex bound to DNA. View the interactive [http://www.rcsb.org/pdb/static.do?p=explorer/viewers/jmol.jsp Jmol image] (PDB 1ZR4).]]'''Biological System''': The biological representation of a pancake is a functional unit of DNA such as a promoter or coding region. To flip these units of DNA, we have reconstituted the Hin/ Hix invertase system (Figure 2) from ''Salmonella typhimurium'' as a BioBrick compatible system in ''E. coli''. '''Hin invertase''' (<partinfo>J31001</partinfo>) was cloned from ''S. typhimurium'', Ames strain TA100 and tagged with LVA. We built the recombinational enhancer (RE) and Hin invertase recognition sequence HixC using the publicly available genomic sequence of ''S. typhimurium'' and [http://gcat.davidson.edu/IGEM06/oligo.html a dsDNA assembly program we created] for gene synthesis from overlapping oligos. | |[[Image:Jmol_Hin_tetrad_DNA.gif|thumb|250px|right|'''Figure 2''' 3-D structure of a Hin protein complex bound to DNA. View the interactive [http://www.rcsb.org/pdb/static.do?p=explorer/viewers/jmol.jsp Jmol image] (PDB 1ZR4).]]'''Biological System''': The biological representation of a pancake is a functional unit of DNA such as a promoter or coding region. To flip these units of DNA, we have reconstituted the Hin/ Hix invertase system (Figure 2) from ''Salmonella typhimurium'' as a BioBrick compatible system in ''E. coli''. '''Hin invertase''' (<partinfo>J31001</partinfo>) was cloned from ''S. typhimurium'', Ames strain TA100 and tagged with LVA. We built the recombinational enhancer (RE) and Hin invertase recognition sequence HixC using the publicly available genomic sequence of ''S. typhimurium'' and [http://gcat.davidson.edu/IGEM06/oligo.html a dsDNA assembly program we created] for gene synthesis from overlapping oligos. | ||
| - | Every segment of DNA flanked by a pair of HixC sites is a "pancake" capable of being inverted. Hin invertase recognizes pairs of HixC sites and inverts the DNA fragment in between the two HixC sites with the help of the Fis protein bound to the RE. In our system, selectable phenotypes (including antibiotic resistance and RFP expression), depend upon the proper arrangement of a series of HixC-flanked DNA segments in a plasmid. This allows us to select for cells that have successfully solved the puzzle. An example of | + | Every segment of DNA flanked by a pair of HixC sites is a "pancake" capable of being inverted. Hin invertase recognizes pairs of HixC sites and inverts the DNA fragment in between the two HixC sites with the help of the Fis protein bound to the RE. In our system, selectable phenotypes (including antibiotic resistance and RFP expression), depend upon the proper arrangement of a series of HixC-flanked DNA segments in a plasmid. This allows us to select for cells that have successfully solved the puzzle. An example of a sorted stack of two pancakes is shown in Figure 3. |
|- valign="top" | |- valign="top" | ||
| - | |[[Image:biocake.jpg|thumb|250px|left|'''Figure 3''' | + | |[[Image:biocake.jpg|thumb|250px|left|'''Figure 3''' Encoding a sorted stack of two biological pancakes]] |
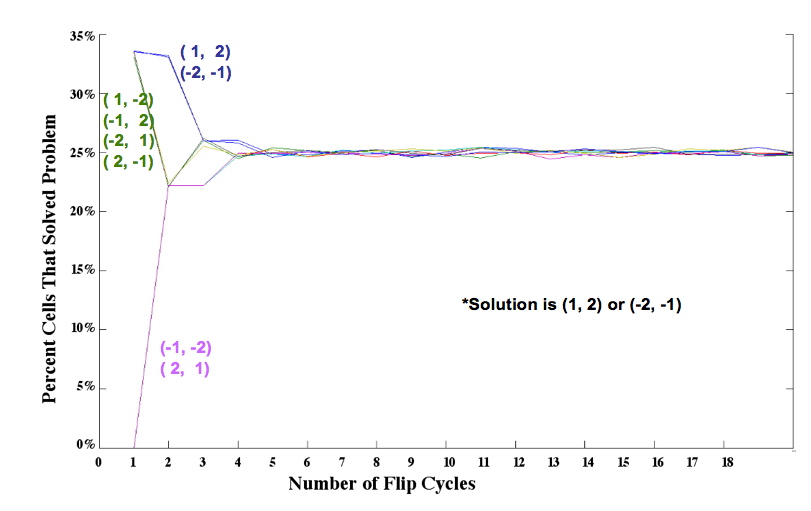
A population of E. coli cells (10<sup>15</sup> cells, for instance) each carrying ~100 copies of pancake stacks has astounding parallel processing capacity. | A population of E. coli cells (10<sup>15</sup> cells, for instance) each carrying ~100 copies of pancake stacks has astounding parallel processing capacity. | ||
Revision as of 17:35, 28 October 2006
 Project Overview [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2006&group=Davidson Davidson Parts] Team Members Tools and Resources Check out our [http://www.bio.davidson.edu/courses/synthetic/photos/FlapJack_HotCakes.html Official Team Photo] |
Our goal is to genetically engineer a biological system that can compute solutions to a puzzle called the burnt pancake problem. The EHOP computer is a proof of concept for computing in vivo, with implications for future data storage devices and transgenic systems. Our work was done in collaboration with the [http://2006.igem.org/wiki/index.php/Missouri_Western_State_University_2006 Missouri Western iGEM Team] and an undergraduate research fellow from [http://www.hamptonu.edu/ Hampton University].
The Burnt Pancake Problem
The [http://en.wikipedia.org/wiki/Pancake_sorting pancake problem] is a puzzle in which a scrambled series of units (or stack of pancakes) must be shuffled into the correct order and orientation. You can try solving [http://www.cut-the-knot.org/SimpleGames/Flipper.shtml a simple version of the pancake problem] yourself.
| In the burnt pancake problem, each pancake is given an orientation by burning one side. Figure 1 shows a scrambled stack of burnt pancakes. To solve the problem, every unit, or pancake, must be placed in the proper order (largest on bottom, smallest on top) and in the proper orientation (burnt side down, golden side up). The pancakes are flipped with two spatulas: one to lift pancakes off the top of the stack, the other to flip part (or all) of the remaining stack of pancakes. The pancakes lifted by the first spatula are returned to the top of the stack without being flipped. You can watch Media:burnt_pancake.ogg a movie of the stack in Figure 1 being sorted to see how the puzzle is solved.
Building the Biological System
TEAM MEMBERS
Faculty
TOOLS AND RESOURCES
White Board Biology Tools (Wet Bench)
Math Tools
Bio-Math Tools
Assembly Plans Progress |