Swimmy Bacteria : Chiba 2006
From 2006.igem.org
(→Referenses) |
(→Experiments) |
||
| (16 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
'''Control the E.coli's movement with light!''' | '''Control the E.coli's movement with light!''' | ||
| - | = | + | =Design= |
| + | |||
| + | to stop the e.coli by light, we needed a part that stop the e.coli, and combine this with a light receptor that UCSF made it last year. | ||
| + | |||
| + | there was some ways to stop the e.coli running: | ||
| + | #making a chemaera with chemotaxis receptor and light sensing receptor | ||
| + | #stop the motor protein's expression(motB protein: from last year's parts) | ||
| + | #controling chemotaxis | ||
| + | etc | ||
| + | |||
| + | we chose to control chemotaxis. | ||
| + | We've found out, from the paper(..see refrences), the chemoreceptor that fix the e.coli tumble. | ||
| + | it is a fragment of the chemoreceptor "Tsr" (a serin receptor)from aa 290 to 551. we call this tsr-cw(or tsr290). | ||
| + | |||
| + | <br /> | ||
[[Image:pathway.jpg|pathway]] | [[Image:pathway.jpg|pathway]] | ||
| + | |||
| + | =Experiments= | ||
| + | we made the new part rbs-tsrcw by PCR and made a new BioBrick (BBa_J29049). | ||
| + | |||
| + | ==Swarm Plate Assay== | ||
| + | first we wanted to make sure this parts will work, so we put it suffix in the OmpC-promotor& araC-promotor. | ||
| + | we found the OmpC promotor (R0082) was not working good, so then we used the araC promotor. | ||
| + | -- results under construction -- | ||
| + | |||
| + | |||
| + | ==Capillary Assay== | ||
| + | we wanted to see if the tsr-cw is working good with this assay,too. | ||
| + | we're waiting for the results... | ||
=Members= | =Members= | ||
| Line 16: | Line 43: | ||
*[[User:Maiko|Maiko Furubayashi]] | *[[User:Maiko|Maiko Furubayashi]] | ||
| - | = | + | =News= |
| - | *'''10/11''' we're finding the suitable media to observe the E.coli's movement(chemotaxis) | + | ==light cannon== |
| + | *'''10/15''' we wrote a picture at the solid media.(not finished yet) also we changed the arabinose concentration at the liquid media. | ||
| + | *'''10/14''' the Fluorescent lamp was the best light. we could see the on/off clearly in the solid media. we found that the thin media is better than the thicker one. | ||
| + | *'''10/11''' we changed the arabinose & S-gal's concentration & the light sorce to find the best condition. | ||
| + | |||
| + | ==swimmy== | ||
| + | *'''10/15''' tried the capiraly assay to see chemotaxis. also we washed the e.coli and observed it with the microscope(but it was kind of hard) | ||
| + | *'''10/11''' we're finding the suitable media to observe the E.coli's movement(chemotaxis). | ||
*'''8/23''' we made the tsr gene parts by PCR & discussed about the locked Tsr mutants. | *'''8/23''' we made the tsr gene parts by PCR & discussed about the locked Tsr mutants. | ||
=Todo List= | =Todo List= | ||
| - | *[[chiba/swimmy/todo| | + | *[[chiba/swimmy/todo|memo]] |
| + | *[[chiba/swimmy/todo/10/19|10/19]] | ||
| + | *[[chiba/swimmy/todo/10/18|10/18]] | ||
| + | *[[chiba/swimmy/todo/10/16|10/16]] | ||
*[[chiba/swimmy/todo/10/15|10/15]] | *[[chiba/swimmy/todo/10/15|10/15]] | ||
*[[chiba/swimmy/todo/10/14|10/14]] | *[[chiba/swimmy/todo/10/14|10/14]] | ||
| Line 34: | Line 71: | ||
==Experiments== | ==Experiments== | ||
| + | *''Microscope'' -- Real-time imaging of fluorescent flagellar filaments.J.bacteriol.2000 | ||
*''Chemotaxis Assay'' -- '''J.Adler''' -- A Method for Measuring Chemotaxis and Use of the Method to Determine Optimum Conditions for Chemotaxis by Escherichia coli | *''Chemotaxis Assay'' -- '''J.Adler''' -- A Method for Measuring Chemotaxis and Use of the Method to Determine Optimum Conditions for Chemotaxis by Escherichia coli | ||
*'''M.K.Slocum and J.S.Parkinson''' (1985) -- Genetics of Methyl-Accepting Chemotaxis Proteins in Escherichia coli: Null Phenotypes of the tar and tap Genes (J.Bacteriol.) | *'''M.K.Slocum and J.S.Parkinson''' (1985) -- Genetics of Methyl-Accepting Chemotaxis Proteins in Escherichia coli: Null Phenotypes of the tar and tap Genes (J.Bacteriol.) | ||
| Line 39: | Line 77: | ||
=Last Update= | =Last Update= | ||
| + | *'''10/13''' todo list | ||
*'''10/11''' swimmy & gene pathway | *'''10/11''' swimmy & gene pathway | ||
*'''8/23''' refrences | *'''8/23''' refrences | ||
*'''8/7''' concept&members --maiko | *'''8/7''' concept&members --maiko | ||
| + | |||
| + | |||
| + | =note= | ||
| + | *[[chiba/note/1|止まる大腸菌について]] | ||
Latest revision as of 16:10, 29 October 2006
Contents |
Concept
Swimmy Bacteria -- the E.coli freeze and gather when they are caught by red light.


red light on -> stop
red light off -> move
Control the E.coli's movement with light!
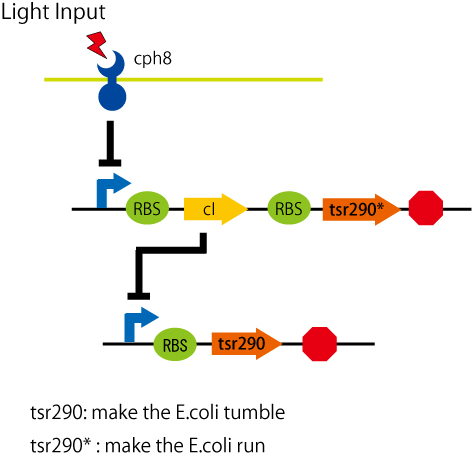
Design
to stop the e.coli by light, we needed a part that stop the e.coli, and combine this with a light receptor that UCSF made it last year.
there was some ways to stop the e.coli running:
- making a chemaera with chemotaxis receptor and light sensing receptor
- stop the motor protein's expression(motB protein: from last year's parts)
- controling chemotaxis
etc
we chose to control chemotaxis. We've found out, from the paper(..see refrences), the chemoreceptor that fix the e.coli tumble. it is a fragment of the chemoreceptor "Tsr" (a serin receptor)from aa 290 to 551. we call this tsr-cw(or tsr290).
Experiments
we made the new part rbs-tsrcw by PCR and made a new BioBrick (BBa_J29049).
Swarm Plate Assay
first we wanted to make sure this parts will work, so we put it suffix in the OmpC-promotor& araC-promotor. we found the OmpC promotor (R0082) was not working good, so then we used the araC promotor. -- results under construction --
Capillary Assay
we wanted to see if the tsr-cw is working good with this assay,too. we're waiting for the results...
Members
Chiba 2006 team あ
News
light cannon
- 10/15 we wrote a picture at the solid media.(not finished yet) also we changed the arabinose concentration at the liquid media.
- 10/14 the Fluorescent lamp was the best light. we could see the on/off clearly in the solid media. we found that the thin media is better than the thicker one.
- 10/11 we changed the arabinose & S-gal's concentration & the light sorce to find the best condition.
swimmy
- 10/15 tried the capiraly assay to see chemotaxis. also we washed the e.coli and observed it with the microscope(but it was kind of hard)
- 10/11 we're finding the suitable media to observe the E.coli's movement(chemotaxis).
- 8/23 we made the tsr gene parts by PCR & discussed about the locked Tsr mutants.
Todo List
Referenses
Locked Tsr Mutants
- A.Bren and M.Eisenbach (2000) -- How Signals Are Heard during Bacterial Chemotaxis: Protein-Protein Interactions in Sensory Signal Propagation (J.Bacteriol.)
- P.Ames and J.S.Parkinson (1988) -- Transmembrane Signaling by Bacterial Chemoreceptors: E.coli Transducers with Locked Signal Output (Cell)
- P.Ames and J.S.Parkinson (1994) -- Constitutively Signaling Fragments of Tsr, the Escherichia coli Serine Chemoreceptor (J.Bacteriol.)
Experiments
- Microscope -- Real-time imaging of fluorescent flagellar filaments.J.bacteriol.2000
- Chemotaxis Assay -- J.Adler -- A Method for Measuring Chemotaxis and Use of the Method to Determine Optimum Conditions for Chemotaxis by Escherichia coli
- M.K.Slocum and J.S.Parkinson (1985) -- Genetics of Methyl-Accepting Chemotaxis Proteins in Escherichia coli: Null Phenotypes of the tar and tap Genes (J.Bacteriol.)
- J.S.Parkinson (1976) --- cheA, cheB, and cheC of Escherichia coli and Their Role in Chemotaxis (J.Bacteriol.)
Last Update
- 10/13 todo list
- 10/11 swimmy & gene pathway
- 8/23 refrences
- 8/7 concept&members --maiko