4-State Device
From 2006.igem.org
(Added pics on assembly, tabels and parts of text.) |
|||
| Line 1: | Line 1: | ||
Back to the [[ETH Zurich]] main page. | Back to the [[ETH Zurich]] main page. | ||
| - | =Organisation= | + | = Organisation = |
| - | ===Group Members=== | + | === Group Members === |
*[[Alexander Roth]](coordinator), [[User:realUACM|Urs A. Müller]], [[Simon Barkow]], [[User:Tamara|Tamara Ulrich]], [[Robin Künzler]], Herve, Dominic, Christophe ... | *[[Alexander Roth]](coordinator), [[User:realUACM|Urs A. Müller]], [[Simon Barkow]], [[User:Tamara|Tamara Ulrich]], [[Robin Künzler]], Herve, Dominic, Christophe ... | ||
| - | ===Meetings=== | + | === Meetings === |
'''2005.08.17, Wednesday, 15:30 @ Alex' bureau (CAB F62)''' [[Summary]] | '''2005.08.17, Wednesday, 15:30 @ Alex' bureau (CAB F62)''' [[Summary]] | ||
'''2005.08.18, Thursday, 13:15 @ Alex' bureau (CAB F62)''' | '''2005.08.18, Thursday, 13:15 @ Alex' bureau (CAB F62)''' | ||
| Line 15: | Line 15: | ||
'''2005.09.06, Tuesday 08:00 @ Alex' bureau (CAB F62)''' | '''2005.09.06, Tuesday 08:00 @ Alex' bureau (CAB F62)''' | ||
| - | =Description of NOR module= | + | = Description of the NOR module = |
This module behaves like a NOR gate. It has two inputs and one output. The output is high only when neither input 1 nor input 2 is high. | This module behaves like a NOR gate. It has two inputs and one output. The output is high only when neither input 1 nor input 2 is high. | ||
| - | ==NOR | + | == NOR Gate == |
| - | + | In the figure below is a parts-view of the NOR gate module with PoPS interfaces (i/o). | |
--------------------------------- | --------------------------------- | ||
| Line 33: | Line 33: | ||
--------------------------------- | --------------------------------- | ||
| - | = | + | == Interconnected NOR module == |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | = Design = | |
| - | == | + | == Repressors == |
| - | + | The Interconnected NOR module consists of four repressors. We considered several alternatives but finally decided to put all out bets on a approach based on zinc finger proteins. | |
| - | We | + | |
| - | + | RNA polymerase... | |
| - | + | === Zinc Finger Proteins === | |
| + | A Zinc Finger (ZF) [http://en.wikipedia.org/wiki/Zinc_finger] is a protein domain [http://en.wikipedia.org/wiki/Protein_domain] that binds to three base pairs of double stranded DNA. A Zinc Finger Protein (ZFP) consists of one or several zinc finger domains. Many protein-DNA interaction for ZF domains and triplet of base pairs have been described, therefore making it possible to to construct artificial transcription factors by combining ZF domains in a modular fashion. The idea is to use a ZFP as a repressor by putting a binding site for a ZFP upstream of the coding region and thereby preventing RNA polymerase to transcribe the gene. | ||
| - | + | We use several different designs of ZFPs. | |
| - | + | ||
| - | + | - Three ZF domains to recognize nine base pairs. | |
| - | + | - Two ZF domains and a leucine zipper domain. The leucine zipper will be cause the protein to home-dimerize and hence it will recognize 12 base pairs. | |
| - | + | - Six ZF domains to recognize twelve base pairs. | |
| - | + | - Three ZF domains fused to negative transcription factors. (e.g. NTD and ERD) | |
| - | |||
| + | ===== Zinc Finger Protein Binding Sites ===== | ||
| + | Since we are going to synthesize parts of our module, we have the possibility to design our own ZFP binding sites, as well as our ZFPs. | ||
| + | We have two alternatives for placing the binding sites. | ||
| - | + | 1. Binding sites in the promoter. This would prevent the polymerase from binding to the promotor. Although this might be most likely to work, we have chosen to not pursue this alternative, while we are quite intrigued by the possibility of a roadblock. | |
| - | {| border="1" | + | 2. Binding sites directly after the start of transcription and before the ribosome binding site. This alternative is attractive, since it would allow for a high degree of modularity. In theory the ZFP would act as an extra "roadblock-operator" and any promotor could be used in front of the protein. |
| - | ! || | + | |
| - | |- | + | The ZFP roadblock operator regions (from now on refered to as: operators) consists of binding motifs for two repressors. The two bindning motifs are usually spaced with 5 base pairs (gcgcg). Some data of the binding strength were available and we have chosen operators with the affinity estimated to K_d = 3-40 nM. |
| - | + | ||
| - | | | + | {| border="1" cellpadding="2" |
| + | ! Repressor !! Binding motif !! K_d (nM) !! freq. in genome !! freq. hamming dist=1 | ||
| + | |- | ||
| + | | R1 || ggaggggac || 4 || 1 || 132 | ||
| + | |- | ||
| + | | R2 || ggaggcggg || 30 || 5 || 396 | ||
| + | |- | ||
| + | | R3 || gggggcgag || 3 || 8 || 303 | ||
| + | |- | ||
| + | | R4 || ggggccgga || 45 || 23 || 433 | ||
| + | |- | ||
| + | | R1 lz || gtccccggggac || N/A || 0 || 2 | ||
|- | |- | ||
| - | | | + | | R2 lz || ctcgccggcgag || N/A || 0 || 7 |
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | | R3 lz || cccgccggcggg || N/A || 0 || 19 |
|- | |- | ||
| - | + | | R4 lz || tccggcgccgga || N/A || 0 || 27 | |
|} | |} | ||
| + | We need to have 4 operators = 4 permutations of ZFP. | ||
| + | The operator regions (i.e. the "roadblocks" that will prevent of the RNApolymerase to transcribe the gene) form a BioBrick that should be inserted between the promoter region and the ribosome binding site in order to keep the design modular. | ||
| - | ====Repression==== | + | === Tester for Zinc Finger === |
| + | To test whether our assumptions about using multiple zinc finger proteins (ZFP) as repressors (i.e. roadblocks) will actually work, we will build a tester/debugging device in parallel with the counter. | ||
| + | |||
| + | === Interconnected NOR System === | ||
| + | The NOR system consists of 4 proteins with 4 operator regions. It has an interface boundary with input module (Pr and Prm). Repressor R3 is connected to a reporter to be able to count to modulo two. | ||
| + | |||
| + | |||
| + | |||
| + | ==== Repression ==== | ||
Note that in both cases, it is still under discussion as whether the repression domain should really be included or not. If it turns out that repression has been used in all the literature we (well, actually Hervé) can find, then we would be better off to play safe and include them: | Note that in both cases, it is still under discussion as whether the repression domain should really be included or not. If it turns out that repression has been used in all the literature we (well, actually Hervé) can find, then we would be better off to play safe and include them: | ||
* Beerli PNAS 1998: they fused ZF to KRAB repressor and it has a stronger effect compare to ZF alone (see p14632 graph A) | * Beerli PNAS 1998: they fused ZF to KRAB repressor and it has a stronger effect compare to ZF alone (see p14632 graph A) | ||
| Line 93: | Line 107: | ||
An alternative could be a negative transcription domain fused to the ZFP. | An alternative could be a negative transcription domain fused to the ZFP. | ||
| - | ====Degradation==== | + | ==== Degradation ==== |
We considered adding a variant with degradation tag, since the system might be slow otherwise. However, due to the fact that we have limited resources - in terms of money, manpower, and time - and the fact that the degradation is decoupled and it will be clearly visible if the system turns out to be too slow, we decided against it for now. | We considered adding a variant with degradation tag, since the system might be slow otherwise. However, due to the fact that we have limited resources - in terms of money, manpower, and time - and the fact that the degradation is decoupled and it will be clearly visible if the system turns out to be too slow, we decided against it for now. | ||
| - | ====Additional Comments==== | + | ==== Additional Comments ==== |
We changed our strategy a couple of times in heated debates. There is quite obviously a trade-off between probability of success through redundancy, i.e. trying multiple approaches in parallel, and limited resources, i.e. financial, manpower, and time. Thus we decided against certain additional variants and cloning steps since we simply won't have the time for it. | We changed our strategy a couple of times in heated debates. There is quite obviously a trade-off between probability of success through redundancy, i.e. trying multiple approaches in parallel, and limited resources, i.e. financial, manpower, and time. Thus we decided against certain additional variants and cloning steps since we simply won't have the time for it. | ||
Another important aspect is the overall goal of keeping the design modular - one of the most important aspects of the this contest. | Another important aspect is the overall goal of keeping the design modular - one of the most important aspects of the this contest. | ||
The modification of the promoter regions of the lambda-system, i.e. Pr and Prm, although a valid approach will probably be dropped, since it is too much work and cost and no longer modular (as opposed to having the operators after the promoter). | The modification of the promoter regions of the lambda-system, i.e. Pr and Prm, although a valid approach will probably be dropped, since it is too much work and cost and no longer modular (as opposed to having the operators after the promoter). | ||
| - | ===Assembly=== | + | There has been some discussion about having the operator around 0 (-9/+1) |
| + | |||
| + | = Synthesis of sequences = | ||
| + | |||
| + | {| border="1" cellpadding="2" | ||
| + | |+Multiplication table | ||
| + | ! Seq nickname !! Length !! Cost !! SacI !! KpnI !! NheI !! EcoRI !! XbaI !! SpeI !! PstI | ||
| + | |- | ||
| + | | J05211+J05100 || 460 || $736.00 || 1 || 74 || 455 || 7,80 || 22,95 || 54,435 || 68,449 | ||
| + | |- | ||
| + | | J05212+J05102 || 460 || $736.00 || 1 || 74 || 455 || 7,80 || 22,95 || 54,435 || 68,449 | ||
| + | |- | ||
| + | | J05213+J05101 || 460 || $736.00 || 1 || 74 || 455 || 7,80 || 22,95 || 54,435 || 68,449 | ||
| + | |- | ||
| + | | J05214+J05103 || 460 || $736.00 || 1 || 74 || 455 || 7,80 || 22,95 || 54,435 || 68,449 | ||
| + | |- | ||
| + | | J05218+J05108 || 515 || $824.00 || 1 || 81 || 510 || 7,87 || 22,102|| 61,490 || 75,504 | ||
| + | |- | ||
| + | | J05109 || 414 || $662.40 || 1 || 409 ||- || 7 || 22 || 389 || 403 | ||
| + | |- | ||
| + | | J05222+J05115 || 676 || $1081.60 || 1 || 87 ||671 || 7,93 || 22,108|| 67,651 || 81,665 | ||
| + | |- | ||
| + | | J05221+J05114 || 728 || $1164.80 || 1 || 133 || 723 || 7,139 || 22,154|| 113,703|| 127,717 | ||
| + | |- | ||
| + | | J05112 || 524 || $838.40 || 1 || 519 || - || 7 || 7 || 499 || 513 | ||
| + | |} | ||
| + | |||
| + | = Assembly = | ||
| + | |||
| + | == ZFP Tester == | ||
* Cut each system into BioBricks Pieces using different restriction enzymes. | * Cut each system into BioBricks Pieces using different restriction enzymes. | ||
* Assemble in parallel for each genes 1-4 in two variants: promoter+operator, rbs+gene | * Assemble in parallel for each genes 1-4 in two variants: promoter+operator, rbs+gene | ||
| Line 107: | Line 150: | ||
* ... | * ... | ||
| - | [[Image: | + | [[Image:TesterParts.gif]] |
| - | ( | + | ==== Tests ==== |
| + | {| | ||
| + | | Repressor || Operator || Comments | ||
| + | |- | ||
| + | | J05100 || J05212 || ... | ||
| + | |- | ||
| + | | J05100 || J05212 || ... | ||
| + | |} | ||
| + | (complete this...) | ||
| + | |||
| + | == Interconnected NOR device == | ||
| + | |||
| + | [[Image:CounterParts.gif]] | ||
=References= | =References= | ||
| Line 143: | Line 198: | ||
[[Newman03]] ("Leucine Zippers") | [[Newman03]] ("Leucine Zippers") | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 15:35, 22 September 2005
Back to the ETH Zurich main page.
Contents |
Organisation
Group Members
- Alexander Roth(coordinator), Urs A. Müller, Simon Barkow, Tamara Ulrich, Robin Künzler, Herve, Dominic, Christophe ...
Meetings
2005.08.17, Wednesday, 15:30 @ Alex' bureau (CAB F62) Summary 2005.08.18, Thursday, 13:15 @ Alex' bureau (CAB F62) 2005.08.22, Monday, 13:45 @ Alex' bureau (CAB F62) Summary 050822 2005.08.25, Thursday 15:30 @ Alex' bureau (CAB F62) 2005.08.29, Monday 14:00 @ Alex' bureau (CAB F62) 2005.08.30, Tuesday 16:00 @ Alex' bureau (CAB F62) 2005.09.06, Tuesday 08:00 @ Alex' bureau (CAB F62)
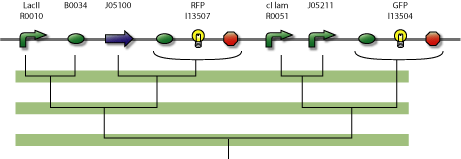
Description of the NOR module
This module behaves like a NOR gate. It has two inputs and one output. The output is high only when neither input 1 nor input 2 is high.
NOR Gate
In the figure below is a parts-view of the NOR gate module with PoPS interfaces (i/o).
---------------------------------
| ------------\ |
PoPS_in1 ---->| | Repressor1 | -------- |
| ------------/ | |
| = |
| --------|------> PoPS_out
| = |
| ------------\ | |
PoPS_in2 ---->| | Repressor2 | -------- |
| ------------/ |
---------------------------------
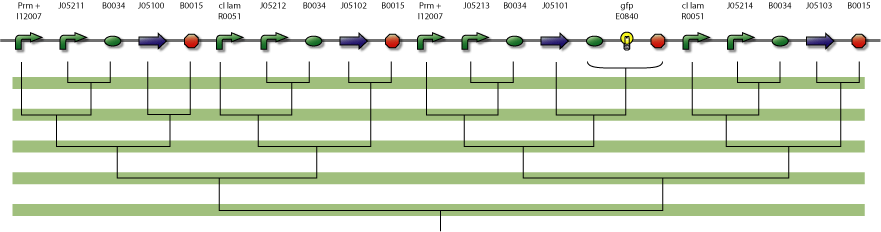
Interconnected NOR module
Design
Repressors
The Interconnected NOR module consists of four repressors. We considered several alternatives but finally decided to put all out bets on a approach based on zinc finger proteins.
RNA polymerase...
Zinc Finger Proteins
A Zinc Finger (ZF) [http://en.wikipedia.org/wiki/Zinc_finger] is a protein domain [http://en.wikipedia.org/wiki/Protein_domain] that binds to three base pairs of double stranded DNA. A Zinc Finger Protein (ZFP) consists of one or several zinc finger domains. Many protein-DNA interaction for ZF domains and triplet of base pairs have been described, therefore making it possible to to construct artificial transcription factors by combining ZF domains in a modular fashion. The idea is to use a ZFP as a repressor by putting a binding site for a ZFP upstream of the coding region and thereby preventing RNA polymerase to transcribe the gene.
We use several different designs of ZFPs.
- Three ZF domains to recognize nine base pairs.
- Two ZF domains and a leucine zipper domain. The leucine zipper will be cause the protein to home-dimerize and hence it will recognize 12 base pairs.
- Six ZF domains to recognize twelve base pairs.
- Three ZF domains fused to negative transcription factors. (e.g. NTD and ERD)
Zinc Finger Protein Binding Sites
Since we are going to synthesize parts of our module, we have the possibility to design our own ZFP binding sites, as well as our ZFPs. We have two alternatives for placing the binding sites.
1. Binding sites in the promoter. This would prevent the polymerase from binding to the promotor. Although this might be most likely to work, we have chosen to not pursue this alternative, while we are quite intrigued by the possibility of a roadblock.
2. Binding sites directly after the start of transcription and before the ribosome binding site. This alternative is attractive, since it would allow for a high degree of modularity. In theory the ZFP would act as an extra "roadblock-operator" and any promotor could be used in front of the protein.
The ZFP roadblock operator regions (from now on refered to as: operators) consists of binding motifs for two repressors. The two bindning motifs are usually spaced with 5 base pairs (gcgcg). Some data of the binding strength were available and we have chosen operators with the affinity estimated to K_d = 3-40 nM.
| Repressor | Binding motif | K_d (nM) | freq. in genome | freq. hamming dist=1 |
|---|---|---|---|---|
| R1 | ggaggggac | 4 | 1 | 132 |
| R2 | ggaggcggg | 30 | 5 | 396 |
| R3 | gggggcgag | 3 | 8 | 303 |
| R4 | ggggccgga | 45 | 23 | 433 |
| R1 lz | gtccccggggac | N/A | 0 | 2 |
| R2 lz | ctcgccggcgag | N/A | 0 | 7 |
| R3 lz | cccgccggcggg | N/A | 0 | 19 |
| R4 lz | tccggcgccgga | N/A | 0 | 27 |
We need to have 4 operators = 4 permutations of ZFP. The operator regions (i.e. the "roadblocks" that will prevent of the RNApolymerase to transcribe the gene) form a BioBrick that should be inserted between the promoter region and the ribosome binding site in order to keep the design modular.
Tester for Zinc Finger
To test whether our assumptions about using multiple zinc finger proteins (ZFP) as repressors (i.e. roadblocks) will actually work, we will build a tester/debugging device in parallel with the counter.
Interconnected NOR System
The NOR system consists of 4 proteins with 4 operator regions. It has an interface boundary with input module (Pr and Prm). Repressor R3 is connected to a reporter to be able to count to modulo two.
Repression
Note that in both cases, it is still under discussion as whether the repression domain should really be included or not. If it turns out that repression has been used in all the literature we (well, actually Hervé) can find, then we would be better off to play safe and include them:
- Beerli PNAS 1998: they fused ZF to KRAB repressor and it has a stronger effect compare to ZF alone (see p14632 graph A)
- Beerli Nat Biotech review feb2002 : if you read the complete paragraph on gene repression (p 132), it gives strong evidence that we should fuse the ZF to a repressor (apparently at the N-Term of the ZF) (either KRAB or SID repressor). It is stated that polymerase blockade through ZF only is not very efficient.
However, the discussion is ongoing: As Alex pointed out we can not be sure that the 45 amino acid long KRAB domain will work, if it don't exists in prokaryotes. KRAB domains have a very specific interaction with a co-repressor molecule. It might be better to find something already known to work in bacteria or skip the repressor approach altogether. An alternative could be a negative transcription domain fused to the ZFP.
Degradation
We considered adding a variant with degradation tag, since the system might be slow otherwise. However, due to the fact that we have limited resources - in terms of money, manpower, and time - and the fact that the degradation is decoupled and it will be clearly visible if the system turns out to be too slow, we decided against it for now.
Additional Comments
We changed our strategy a couple of times in heated debates. There is quite obviously a trade-off between probability of success through redundancy, i.e. trying multiple approaches in parallel, and limited resources, i.e. financial, manpower, and time. Thus we decided against certain additional variants and cloning steps since we simply won't have the time for it. Another important aspect is the overall goal of keeping the design modular - one of the most important aspects of the this contest. The modification of the promoter regions of the lambda-system, i.e. Pr and Prm, although a valid approach will probably be dropped, since it is too much work and cost and no longer modular (as opposed to having the operators after the promoter).
There has been some discussion about having the operator around 0 (-9/+1)
Synthesis of sequences
| Seq nickname | Length | Cost | SacI | KpnI | NheI | EcoRI | XbaI | SpeI | PstI |
|---|---|---|---|---|---|---|---|---|---|
| J05211+J05100 | 460 | $736.00 | 1 | 74 | 455 | 7,80 | 22,95 | 54,435 | 68,449 |
| J05212+J05102 | 460 | $736.00 | 1 | 74 | 455 | 7,80 | 22,95 | 54,435 | 68,449 |
| J05213+J05101 | 460 | $736.00 | 1 | 74 | 455 | 7,80 | 22,95 | 54,435 | 68,449 |
| J05214+J05103 | 460 | $736.00 | 1 | 74 | 455 | 7,80 | 22,95 | 54,435 | 68,449 |
| J05218+J05108 | 515 | $824.00 | 1 | 81 | 510 | 7,87 | 22,102 | 61,490 | 75,504 |
| J05109 | 414 | $662.40 | 1 | 409 | - | 7 | 22 | 389 | 403 |
| J05222+J05115 | 676 | $1081.60 | 1 | 87 | 671 | 7,93 | 22,108 | 67,651 | 81,665 |
| J05221+J05114 | 728 | $1164.80 | 1 | 133 | 723 | 7,139 | 22,154 | 113,703 | 127,717 |
| J05112 | 524 | $838.40 | 1 | 519 | - | 7 | 7 | 499 | 513 |
Assembly
ZFP Tester
- Cut each system into BioBricks Pieces using different restriction enzymes.
- Assemble in parallel for each genes 1-4 in two variants: promoter+operator, rbs+gene
- Assemble the two constructs
- ...
Tests
| Repressor | Operator | Comments |
| J05100 | J05212 | ... |
| J05100 | J05212 | ... |
(complete this...)
Interconnected NOR device
References
Kim & Wang (??)
Park et al. 2005 (Activation of transcription)
Chou et al. 1998
Beerli00 (Linker)
Segal99 ("GNN-paper")
Dreier01 ("ANN-paper")
Dreier05 ("CNN-paper")
Yang95 (Kinetics!!)
Klug05 (Minireview)
Newman03 ("Leucine Zippers")