PROJECT PROPOSAL
From 2006.igem.org
INTRODUCTION
The objective of this project is to engineer Escherichia coli which are able to actively pursue and mark or eliminate another bacterial target. This system can be divided into three components: an input element, a processing element, and a response element. The input element will consist of a quorum sensing circuit which would allow specific detection of the bacterial target. The processing element will facilitate the signaling of this input into controlled responses. A number of different response elements can be conceived, to be used separately or in tandem: 1) integration into the chemotactic pathway of E. coli, allowing for directed mobilization towards the target, 2) reporter response at high pheromone concentrations to allow for visual identification of the target location (e.g., GFP production), and 3) an elimination response to produce molecules which are specifically lethal to the desired target.
SIGNIFICANCE
Successful completion of this project will demonstrate the ability to engineer complex behavioral phenotypes (predatory-prey behavior) into bacterial systems through the integration of biological modules. The phenotype of ‘quorumtaxis’ through the amalgamation of quorum-sensing and chemotaxis pathways would also be a unique demonstration of the novel circuits which can be achieved through the combined power of protein engineering methods and system biology paradigms. Furthermore, progress on this project would improve the tools available for engineering chemotactic responses to non-natural ligands.
In addition, a number of new parts will be made available to the iGEM registry. The most significant of these would be a ComP/Tsr hybrid receptor, which would allow the control of chemotaxis towards concentrations of a peptide pheromone. In addition, if the secondary strategy (outlined in Section III) is undertaken, Tsr-CW and Tsr-CCW receptor fragments would be added to the iGEM registry, allowing for transcriptional control of chemotaxis. The complete ComX quorum-sensing system may also be added to the iGEM registry; this would consist of a 2-gene construct (ComQ, ComX) for the sender device and a 2-gene construct for the receiver (ComP, ComA). Currently, the iGEM registry only contains AHL-based systems of the LuxR receiver family. The addition of a completely different quorum system would greatly expand the tools available for cell-to-cell communication.
RATIONALE
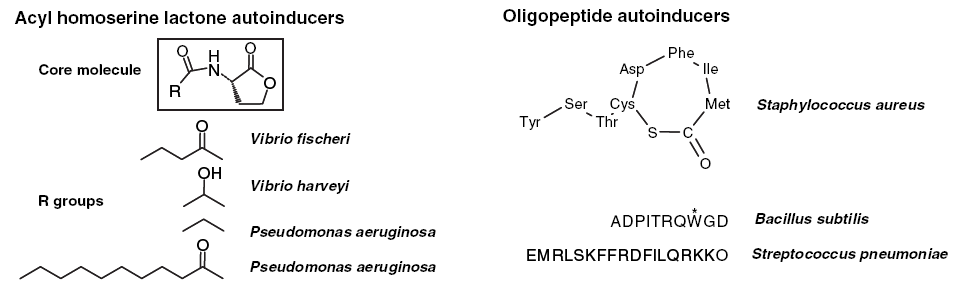
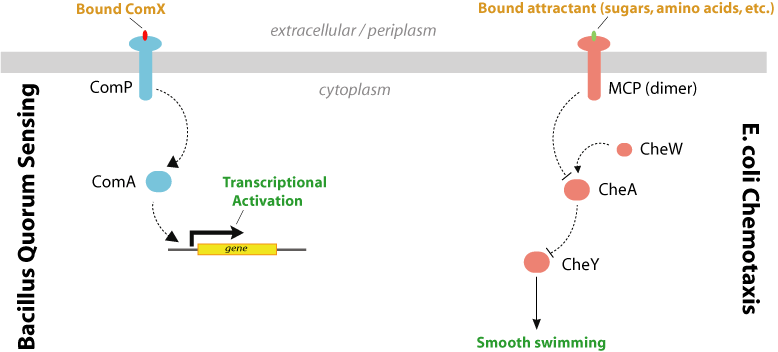
The quorum sensing system of gram negative bacteria is primarily mediated by the production and detection of acylated homoserine lactones (AHL) which accumulate in the extracellular medium as the cellular density increases. In contrast, gram positive bacteria such as Bacillus subtilis use excreted peptide pheromones to achieve a quorum response (Figure 1). Due to the considerable differences between these systems, crosstalk would not be expected to occur. We propose to engineer a response element in E. coli which is able to detect the peptide pheromone of gram positive cells and produce an intracellular signal.
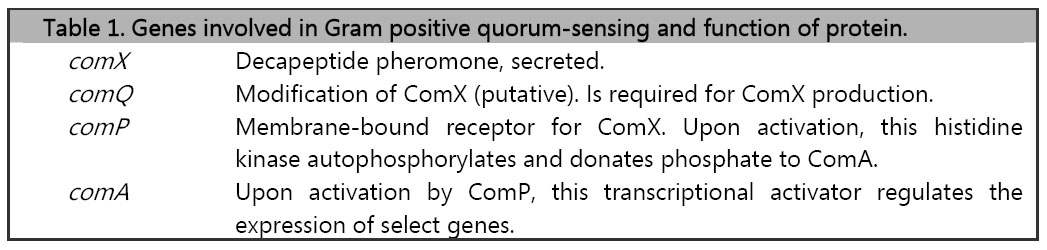
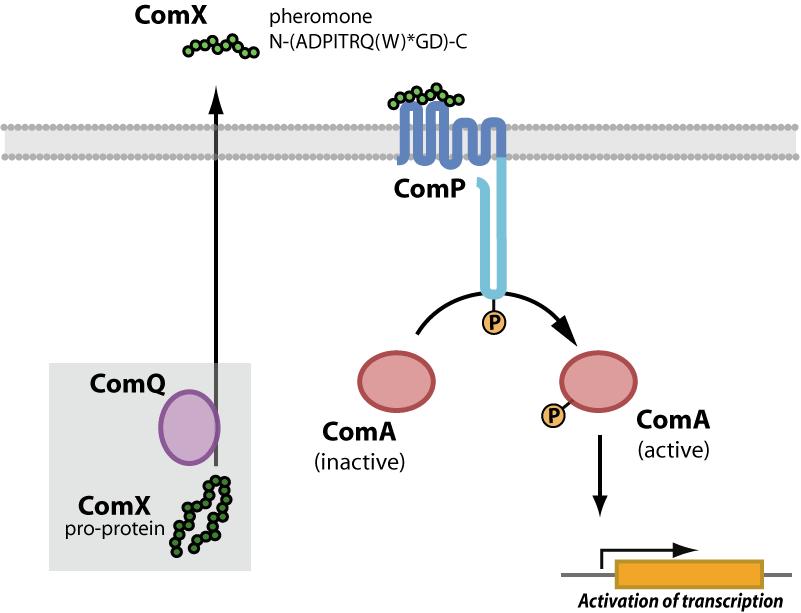
The ComX quorum-sensing system in gram positive bacteria consist of the genes comX, comQ, comP, and comA (Table 1 and Figure 2) which have been shown to regulate genetic competence. These genes cluster occur as an operon in B. subtilis and their protein products have been characterized (1).
The input element will consist of the ComX pheromone, which is a 10-residue peptide secreted extracellularly. In B. subtilis, ComX production requires the presence of comQ. The exact function of ComQ is not well understood. As ComX accumulates extracellularly and detection of cell density is mediated by a ComX receptor, ComP, which is a membrane-bound histidine kinase. ComP autophosphorylates upon binding to ComX and subsequently donates this phosphate to the downstream transcriptional activator, ComA. Once activated, ComA binds to specific binding sites in the regulatory regions of genes involved in genetic competence, antibiotic production, biofilm formation, swarming motility, and membrane secretion (Comella and Grossman, 2005).
A variety of processing elements can be integrated at the genetic level to obtain either simple gradient responses, stable biphasic switches (toggle switch) or integration with another input (AND gates). Many of these modules have been constructed and tested in various iGEM projects. The usage of these various circuits to modulate the system response can be modeled to characterize the dynamics of the genetic circuitry and fine-tuned during empirical testing of the system.
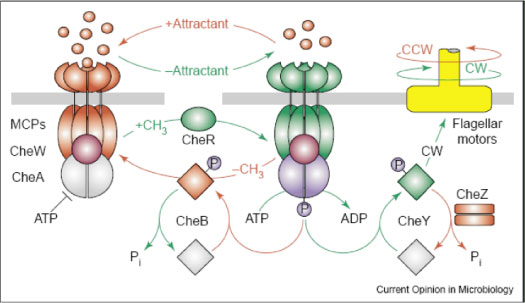
Chemotaxis in bacteria has been extensively studied as a model system for receptor signaling and intracellular signal transduction. To actively seek energy sources or optimal physicochemical environments, bacteria such as E. coli employ a system of alternating tumbling and swimming behaviors. Detection of an extracellular chemical signal produces a swimming phenotype (CCW flagellar rotation); a decrease in the signal induces a tumbling response (CW). The bacterium thus traces out a path similar to a ‘random walk,’ which produces movement in the direction of increasing attractant. Feedback control by methylation of the receptor proteins restores sensitivity to the environmental input by modulating phosphorylation activity. This allows the cell to detect attractant gradients in space as well as time. Figure 3 shows a diagram of chemotaxis from Parkinson, et al., 2005 (2). The chemotaxis system has also been shown to be essential for swarming motility in E. coli(3).
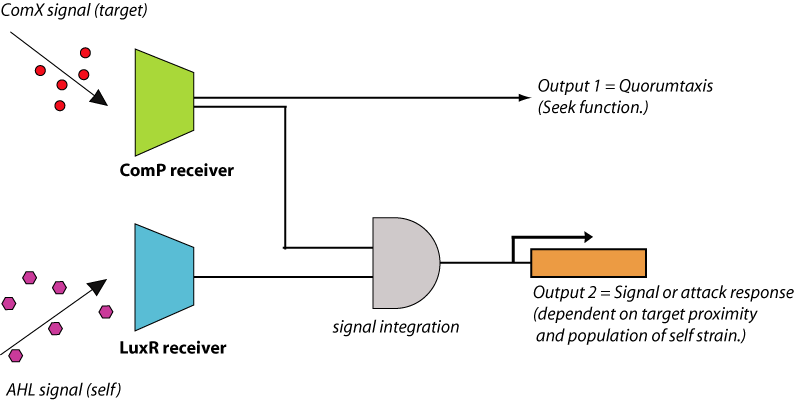
By integrating quorum-sensing and chemotactic pathways, it is possible to engineer a ‘quorumtaxis’ system, whereby a bacterium can sense the quorum pheromone of another species and move in that direction. To date, there have been no published results of engineered quorumtaxis systems. Construction of a functioning quorumtaxis system would be a significant innovation that demonstrates the power of developing a novel complex phenotype from pre-existing bacterial components. This can be done by integration of the ComX receiver system composed of ComP/ComA with the chemotaxis system of E.coli (figure 4). This engineered E. coli will begin to migrate towards gram positive cells (e.g., B. subtilis) due to the presence of extracellular ComX pheromone produced by these cells. Also, by adding the AHL quorum-sensing system to the E. coli cells and integrating the AHL and ComX detection systems together, it would be possible to produce an output which is dependent on both the proximity to the prey cells and the density of the predator strain.
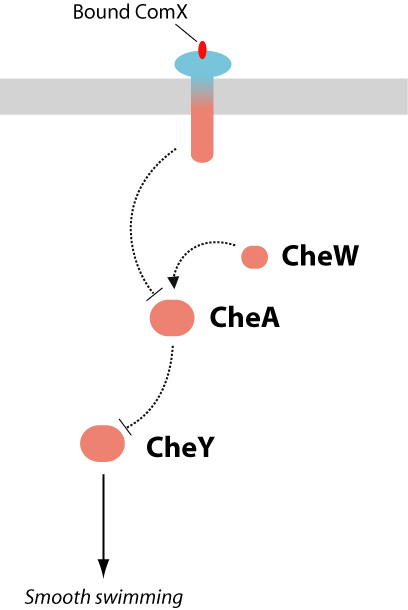
The difficult aspect of this proposed project would be to construct and optimize the quorumtaxis pathway. Flagellar response to the presence of extracellular ComX can be engineered in a number of ways, all of which focus on the integration of signaling pathways between the B. subtilis ComXP and the E. coli chemotaxis system, both shown in Figure 5.
Strategy 1. ComP-Tsr chimeric receptor.
A commonality shared by the ComP-ComA system and the bacterial chemotaxis system is that they are both two-component signaling systems, as shown in Figure 5 (4). In such signaling pathways, an extracellular signal is detected by a membrane receptor, which then activates a signaling cascade, culminating in transcription or phenotypic output. The similarity of the ComP and MCP signaling systems allow for the construction of a hybrid two-component signal-transduction system which contains the extracellular ComP domain (ComX-binding) and the cytoplasmic signaling domains of the chemotaxis receptor Tsr. The engineering of chimeric proteins has been demonstrated for a number of two-component receptors. The modular domain architecture of these receptors has been shown to facilitate swapping of the the extracellular ligand binding domains or the cytoplasmic signaling domains. Table 2 lists a number of successfully engineered hybrid receptors.
Utilizing this tactic in the development of our quorumtactic system, a library of ComP-Tsr chimeras would be produced and tested for the ability to elicit straight swimming in E. coli in the presence of extracellular ComX. On a molecular scale, the desired chimeric protein would be able to bind ComX and possess an allosterically controlled histidine kinase activity (Figure 6). The appropriate fusion protein should consist of 1) the sensor domain of ComP, 2) a transmembrane region, and 3) the cytoplasmic signaling domain of the E. coli chemoreceptor Tsr. This strategy is similar to that employed by the Voigt laboratory, to produce a phytochrome-EnvZ histidine kinase (HK) chimeric protein in which red light allosterically inhibits the autophosphorylation of the HK domain and shuts off gene expression. The advantage of this hybrid-receptor approach is that it retains all the functional elements of the bacterial chemotaxis system in regards to the desensitization adaptation afforded by CheR and CheB-mediated methylation events; the disadvantage is that a major part of the project will involve protein engineering and screening.
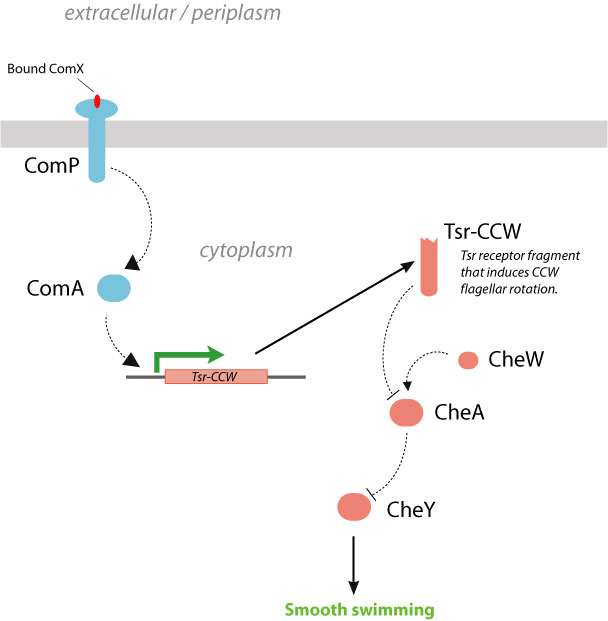
Strategy 2. Expression of Tsr fragments under control of ComA activation. Another alternative approach would be to express an upstream chemotaxis signaling protein under control of ComA. Although there have been no naturally-occuring proteins which can elicit chemotactic responses upon expression (i.e., an internal chemotaxis transcriptional switch), the production of protein fragments have been previously identified with such properties. The serine chemoreceptor Tsr, like most methyl-accepting chemotaxis proteins (MCPs), is a transmembrane protein roughly 550 amino acids in length. It contains a periplasmic ligand-binding domain, a transmembrane domain, and a cytoplasmic signaling domain. Tsr is one of the major chemoreceptors in E. coli and is highly expressed (5). Ames and Parkinson have demonstrated that soluble fragments of the Tsr cytoplasmic signaling domain (residues 290-470) expressed in E. coli Tsr deletion strains (RP5700) are able to alter flagellar rotation (6). Specifically, wildtype Tsr fragments are known to produce a CW signal (tumbling), whereas the same fragment with a single amino-acid mutation A213V shifts the flagellar rotation to CCW (straight swimming). Thus, by placing the expression of Tsr-CCW under the control of the ComA transcriptional activator, it may be possible to integrate the detection of ComX pheromone with smooth forward swimming. However, it is unknown whether these Tsr fragments can be methylated for sensitivity attenuation. In addition, by placing chemotaxis under transcriptional control rather than direct chemical signaling, the response time of the system may be affected.
Signaling/Kill response The engineering of quorumtaxis in E. coli would be the primary task at hand. Construction of downstream functions would be relatively straightforward. For example, if the bacterium is able to detect and swarm towards the gram-positive target cells, we could then design the E. coli to fluoresce (signaling) and/or express antimicrobial peptides (elimination). A number of defensins, such as those produced by certain insects, exhibit bactericidal activity against gram-positive bacteria. Another potential module for a kill response could be the daptomycin biosynthesis gene cluster, which produces a 13 amino acid cyclic lipopeptide with potent activity against gram-positive bacteria. These genes can be engineered with a secretion signal and placed under the control of the ComP-ComA system, with the possibility of integrated control from a quorum-sensing system specifically engineered into the E. coli (e.g., the endogenous AI-2 system or the exogenous AHL system). Thus, these genes will only be expressed under the combined conditions of high levels of ComX, indicating close proximity to gram-positive targets, and a high density of the E. coli itself. Such control ensures that ‘kill response’ is activated at an optimal time to maximize the local concentration of the secreted toxins and minimize adaptation responses from the target cells.
Proposed Experiments
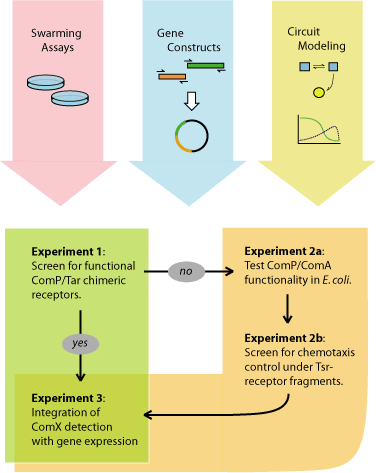
This project can be anticipated to initially proceed in three parallel directions: (1) establishing a working E. coli swarming-plate assay, (2) construction of the ComP/Tar receptor hybrids, and (3) computational modeling of the quorumtaxis genetic elements. The first strategy to be attempted will be the engineering of a functional ComP/Tar chimeric receptor, which will require a functional swarming assay and a library of ComP-Tar gene fusions. If this is unsuccessful, the alternate strategy will be to introduce transcriptional control of the E. coli chemotaxis system by the expression of Tsr receptor fragments. Upon the successful construction of the quorumtaxis system, the downstream output circuits (i.e., pheromone concentration-dependent expression of signaling or toxin genes) will be designed and assembled. Throughout the cloning, screening, and validation of the quorumtaxis system, computational simulations of the genetic regulatory circuits can be used to predict and refine the circuit architecture. In addition, the large-scale dynamics of the phenotype as a predator-prey system can be modeled. Major experimental milestones for this project are shown in Figure 8 and described below.
Experiment 1: Can functional ComP/Tar hybrids be engineered? Using PCR methods, a number of chimeras will be engineered, fusing the ComX-sensing domain of ComP with the cytoplasmic signaling domain of the Tar receptor. Successful mutants will be able to produce a quorumtactic response in the presence of ComX. Swarm screening will be performed in E. coli Tar-deletion mutants.
Experiment 2a: Can the ComP/ComA system be used in E. coli to detect ComX? It is currently unknown whether the ComP/ComA receptor system can function properly in E. coli. This experiment will demonstrate the functionality of the ComP receptor in E.coli. Transformations will be performed of E. coli with vector 1 expressing ComP and ComA, and vector 2 with a reporter gene under control of the ComA transcriptional activation region. Desired phenotypes will be assayed on swarming plates using B. subtilis or artificially produced ComX as bait.
Experiment 2b: Does expression of Tsr fragments allow sufficient control of the E. coli chemotactic pathway? Expression of Tsr fragments known to alter the flagellar rotation of E. coli will be placed under control of the ComP-ComA system. Thus, in the presence of ComX, the appropriate protein will be expressed to control quorumtaxis. Desired phenotypes will be assayed on swarming plates using B. subtilis or artificially produced ComX as bait.
Experiment 3: Can the expression of a gram-positive toxic protein be coupled with the quorumtaxis system? Engineering expression of the toxin will be regulated by the integrated detection of environmental conditions (ComX concentrations and self-density). The regulatory system can be tested by using a reporter protein such as GFP. The expression of GFP can then be followed on swarming plates to determine the efficiency of gene control and expression timing before cloning in the gram-positive toxin gene.
References
1. Tortosa, P., et al. (2001) Journal of Bacteriology 183, 451-460.
2. Parkinson, J. S., Ames, P. & Studdert, C. A. (2005) Curr Opin Microbiol 8, 116-21.
3. Burkart, M., Toguchi, A. & Harshey, R. M. (1998) Proc Natl Acad Sci U S A 95, 2568-73.
4. Weinrauch, Y., Penchev, R., Dubnau, E., Smith, I. & Dubnau, D. (1990) Genes Dev 4, 860-72.
5. Li, M. & Hazelbauer, G. L. (2004) J Bacteriol 186, 3687-94.
6. Ames, P. & Parkinson, J. S. (1994) J Bacteriol 176, 6340-8.